Biology:Chitinase
| Chitinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
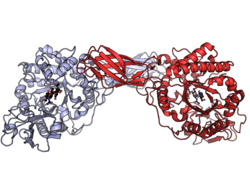 Serratia marcescens chitinase A dimer with bound inhibitor allosamidin. PDB: 1FFQ | |||||||||
| Identifiers | |||||||||
| EC number | 3.2.1.14 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
| chitinase, acidic | |
|---|---|
 | |
| Identifiers | |
| Symbol | CHIA |
| NCBI gene | 27159 |
| HGNC | 17432 |
| OMIM | 606080 |
| RefSeq | NM_001040623 |
| UniProt | Q9BZP6 |
| Other data | |
| Locus | Chr. 1 p13.1-21.3 |
| chitinase 1 (chitotriosidase) | |
|---|---|
 | |
| Identifiers | |
| Symbol | CHIT1 |
| NCBI gene | 1118 |
| HGNC | 1936 |
| OMIM | 600031 |
| RefSeq | NM_003465 |
| UniProt | Q13231 |
| Other data | |
| Locus | Chr. 1 q31-q32 |
Chitinases (EC 3.2.1.14, chitodextrinase, 1,4-β-poly-N-acetylglucosaminidase, poly-β-glucosaminidase, β-1,4-poly-N-acetyl glucosamidinase, poly[1,4-(N-acetyl-β-D-glucosaminide)] glycanohydrolase, (1→4)-2-acetamido-2-deoxy-β-D-glucan glycanohydrolase; systematic name (1→4)-2-acetamido-2-deoxy-β-D-glucan glycanohydrolase) are hydrolytic enzymes that break down glycosidic bonds in chitin.[1] They catalyse the following reaction:
- Random endo-hydrolysis of N-acetyl-β-D-glucosaminide (1→4)-β-linkages in chitin and chitodextrins
As chitin is a component of the cell walls of fungi and exoskeletal elements of some animals (including mollusks and arthropods), chitinases are generally found in organisms that either need to reshape their own chitin[2] or dissolve and digest the chitin of fungi or animals.
Species distribution
Chitinivorous organisms include many bacteria[3] (Aeromonads, Bacillus, Vibrio,[4] among others), which may be pathogenic or detritivorous. They attack living arthropods, zooplankton or fungi or they may degrade the remains of these organisms.
Fungi, such as Coccidioides immitis, also possess degradative chitinases related to their role as detritivores and also to their potential as arthropod pathogens.
Chitinases are also present in plants – for example barley seed chitinase: PDB: 1CNS, EC 3.2.1.14. Barley seeds are found to produce clone 10 in Ignatius et al 1994(a). They find clone 10, a Class I chitinase, in the seed aleurone during development.[5][6][7] Leaves produce several isozymes (as well as several of β-1,3-glucanase). Ignatius et al 1994(b) find these in the leaves, induced by powdery mildew.[5] Ignatius et al also find these (seed and leaf isozymes) to differ from each other.[6][8] Some of these are pathogenesis related (PR) proteins that are induced as part of systemic acquired resistance. Expression is mediated by the NPR1 gene and the salicylic acid pathway, both involved in resistance to fungal and insect attack. Other plant chitinases may be required for creating fungal symbioses.[9]
Although mammals do not produce chitin, they have two functional chitinases, Chitotriosidase (CHIT1) and acidic mammalian chitinase (AMCase), as well as chitinase-like proteins (such as YKL-40) that have high sequence similarity but lack chitinase activity.[10]
Classification
- Endochitinases (EC 3.2.1.14) randomly split chitin at internal sites of the chitin microfibril, forming soluble, low molecular mass multimer products. The multimer products includes di-acetylchitobiose, chitotriose, and chitotetraose, with the dimer being the predominant product.[11]
- Exochitinases have also been divided into two sub categories:
- Chitobiosidases (EC 3.2.1.29) act on the non-reducing end of the chitin microfibril, releasing the dimer, di-acetylchitobiose, one by one from the chitin chain. Therefore, there is no release of monosaccharides or oligosaccharides in this reaction.[12]
- β-1,4- N-acetylglucosaminidases (EC 3.2.1.30) split the multimer products, such as di-acetylchitobiose, chitotriose, and chitotetraose, into monomers of N-acetylglucoseamine (GlcNAc).[11]
Chitinases were also classified based on the amino acid sequences, as that would be more helpful in understanding the evolutionary relationships of these enzymes to each other.[13] Therefore, the chitinases were grouped into three families: 18, 19, and 20.[14] Both families 18 and 19 consists of endochitinases from a variety of different organisms, including viruses, bacteria, fungi, insect, and plants. However, family 19 mainly comprises plant chitinases. Family 20 includes N-acetylglucosaminidase and a similar enzyme, N-acetylhexosaminidase.[13]
And as the gene sequences of the chitinases were known, they were further classified into six classes based on their sequences. Characteristics that determined the classes of chitinases were the N-terminal sequence, localization of the enzyme, isoelectric pH, signal peptide, and inducers.[13]
Class I chitinases had a cysteine-rich N-terminal, leucine- or valine-rich signal peptide, and vacuolar localization. And then, Class I chitinases were further subdivided based on their acidic or basic nature into Class Ia and Class Ib, respectively.[15] Class 1 chitinases were found to comprise only plant chitinases and mostly endochitinases.
Class II chitinases did not have the cysteine-rich N-terminal but had a similar sequence to Class I chitinases. Class II chitinases were found in plants, fungi, and bacteria and mostly consisted of exochitinases.[13]
Class III chitinases did not have similar sequences to chitinases in Class I or Class II.[13]
Class IV chitinases had similar characteristics, including the immunological properties, as Class I chitinases.[13] However, Class IV chitinases were significantly smaller in size compared to Class I chitinases.[16]
Class V and Class VI chitinases are not well characterized. However, one example of a Class V chitinase showed two chitin binding domains in tandem, and based on the gene sequence, the cysteine-rich N-terminal seemed to have been lost during evolution, probably due to less selection pressure that caused the catalytic domain to lose its function.[13]


Function
Like cellulose, chitin is an abundant biopolymer that is relatively resistant to degradation.[17] Many mammals can digest chitin and the specific chitinase levels in vertebrate species are adapted to their feeding behaviours.[18] Certain fish are able to digest chitin.[19] Chitinases have been isolated from the stomachs of mammals, including humans.[20]
Chitinase activity can also be detected in human blood[21][22] and possibly cartilage.[23] As in plant chitinases this may be related to pathogen resistance.[24][25]
Clinical significance
Chitinases production in the human body (known as "human chitinases") may be in response to allergies, and asthma has been linked to enhanced chitinase expression levels.[26][27][28][29][30]
Human chitinases may explain the link between some of the most common allergies (dust mites, mold spores—both of which contain chitin) and worm (helminth) infections, as part of one version of the hygiene hypothesis[31][32][33] (worms have chitinous mouthparts to hold the intestinal wall). Finally, the link between chitinases and salicylic acid in plants is well established[further explanation needed]—but there is a hypothetical link between salicylic acid and allergies in humans.[34][non sequitur]
May be used to monitor enzymotherapy supplementation in Gaucher's disease.[1]
Regulation in fungi
Regulation varies from species to species, and within an organism, chitinases with different physiological functions would be under different regulation mechanisms. For example, chitinases that are involved in maintenance, such as remodeling the cell wall, are constitutively expressed. However, chitinases that have specialized functions, such as degrading exogenous chitin or participating in cell division, need spatio-temporal regulation of the chitinase activity.[35]
The regulation of an endochitinase in Trichoderma atroviride is dependent on a N-acetylglucosaminidase, and the data indicates a feedback-loop where the break down of chitin produces N-acetylglucosamine, which would be possibly taken up and triggers up-regulation of the chitinbiosidases.[36]
In Saccharomyces cerevisiae and the regulation of ScCts1p (S. cerevisiae chitinase 1), one of the chitinases involved in cell separation after cytokinesis by degrading the chitin of the primary septum.[37] As these types of chitinases are important in cell division, there must be tight regulation and activation. Specifically, Cts1 expression has to be activated in daughter cells during late mitosis and the protein has to localize at the daughter site of the septum.[38] And to do this, there must be coordination with other networks controlling the different phases of the cell, such as Cdc14 Early Anaphase Release (FEAR), mitotic exit network (MEN), and regulation of Ace2p (transcription factor) and cellular morphogenesis (RAM)[39] signalling networks. Overall, the integration of the different regulatory networks allows for the cell wall degrading chitinase to function dependent on the cell's stage in the cell cycle and at specific locations among the daughter cells.[35]
Presence in food
Chitinases occur naturally in many common foods. Phaseolus vulgaris,[40] bananas, chestnuts, kiwifruit, avocados, papaya, and tomatoes, for example, all contain significant levels of chitinase, as defense against fungal and invertebrate attack. Stress, or environmental signals like ethylene gas, may stimulate increased production of chitinase.
Some parts of chitinase molecules, almost identical in structure to hevein or other proteins in rubber latex due to their similar function in plant defense, may trigger an allergic cross-reaction known as latex-fruit syndrome.[41]
Applications
Chitinases have a wealth of applications, some of which have already been realized by industry. This includes bio-conversion of chitin to useful products such as fertilizer, the production of non-allergenic, non-toxic, biocompatible, and biodegradable materials (contact lenses, artificial skin and sutures with these qualities are already being produced) and enhancement of insecticides and fungicides.[42] Phaseolus vulgaris chitinase - bean chitinase, BCH - has been transgenically inserted as a pest deterrent into entirely unrelated crops.[40]
Possible future applications of chitinases are as food additives to increase shelf life, therapeutic agent for asthma and chronic rhinosinusitis, as an anti-fungal remedy, an anti-tumor drug and as a general ingredient to be used in protein engineering.[42]
See also
- Ligninase
References
- ↑ Chitin and Chitinases. Basel: Birkhäuser. 1999. ISBN 978-3-7643-5815-0.
- ↑ "Autolysis and aging of Penicillium chrysogenum cultures under carbon starvation: Chitinase production and antifungal effect of allosamidin". The Journal of General and Applied Microbiology 47 (4): 201–211. August 2001. doi:10.2323/jgam.47.201. PMID 12483620.
- ↑ "Chitinase genes in lake sediments of Ardley Island, Antarctica". Applied and Environmental Microbiology 71 (12): 7904–9. December 2005. doi:10.1128/AEM.71.12.7904-7909.2005. PMID 16332766. Bibcode: 2005ApEnM..71.7904X.
- ↑ "Conservation of the chitin utilization pathway in the Vibrionaceae". Applied and Environmental Microbiology 74 (1): 44–51. January 2008. doi:10.1128/AEM.01412-07. PMID 17933912. Bibcode: 2008ApEnM..74...44H.
- ↑ 5.0 5.1 "Pathogenesis-related proteins and their genes in cereals". Plant Cell, Tissue and Organ Culture (Kluwer Academic) 64 (2/3): 93–114. 2001. doi:10.1023/a:1010763506802. ISSN 0167-6857.
- ↑ 6.0 6.1 "Seed chitinases". Seed Science Research (Cambridge University Press (CUP)) 12 (4): 217–230. 2002. doi:10.1079/ssr2002113. ISSN 0960-2585.
- ↑ "Antifungal proteins and other mechanisms in the control of sorghum stalk rot and grain mold". Journal of Agricultural and Food Chemistry (American Chemical Society (ACS)) 49 (10): 4732–4742. October 2001. doi:10.1021/jf010007f. PMID 11600015.
- ↑ Basra, A.S. (2007). Handbook of Seed Science and Technology. Scientific Publishers. pp. 795. doi:10.2307/25065722. ISBN 978-93-88148-36-8. https://books.google.com/books?id=mNaBDwAAQBAJ. Retrieved 2021-11-17. ISBN 9788172335731 ISBN 9388148363.
- ↑ "Differential expression of eight chitinase genes in Medicago truncatula roots during mycorrhiza formation, nodulation, and pathogen infection". Molecular Plant-Microbe Interactions 13 (7): 763–777. July 2000. doi:10.1094/MPMI.2000.13.7.763. PMID 10875337.
- ↑ "Potential role of chitinase 3-like-1 in inflammation-associated carcinogenic changes of epithelial cells". World Journal of Gastroenterology 15 (42): 5249–59. November 2009. doi:10.3748/wjg.15.5249. PMID 19908331.
- ↑ 11.0 11.1 "Chitinases of fungi and plants: their involvement in morphogenesis and host—parasite interaction". FEMS Microbiology Reviews 11 (4): 317–338. 1993-08-01. doi:10.1111/j.1574-6976.1993.tb00004.x.
- ↑ "Chitinolytic Enzymes of Trichoderma harzianum: Purification of Chitobiosidase and Endochitinase". Phytopathology 83 (3): 313. 1993. doi:10.1094/phyto-83-313.
- ↑ 13.0 13.1 13.2 13.3 13.4 13.5 13.6 "Chitinolytic enzymes: an exploration". Enzyme and Microbial Technology 26 (7): 473–483. April 2000. doi:10.1016/s0141-0229(00)00134-4. PMID 10771049.
- ↑ "A classification of glycosyl hydrolases based on amino acid sequence similarities". The Biochemical Journal 280 ( Pt 2) (2): 309–16. December 1991. doi:10.1042/bj2800309. PMID 1747104.
- ↑ "What's new in chitinase research?". Experientia 48 (8): 701–716. August 1992. doi:10.1007/BF02124285. PMID 1516675.
- ↑ "Plant chitinases". The Plant Journal 3 (1): 31–40. January 1993. doi:10.1046/j.1365-313x.1993.t01-1-00999.x. PMID 8401605.
- ↑ "Apparent chitin digestibilities in the Eastern screech owl (Otus asio) and the American kestrel (Falco sparverius)". Journal of Experimental Zoology 283 (4–5): 387–393. 2005. doi:10.1002/(SICI)1097-010X(19990301/01)283:4/5<387::AID-JEZ8>3.0.CO;2-W.
- ↑ "Chitin digestibility is dependent on feeding behaviors, which determine acidic chitinase mRNA levels in mammalian and poultry stomachs". Scientific Reports 8 (1): 1461. January 2018. doi:10.1038/s41598-018-19940-8. PMID 29362395. Bibcode: 2018NatSR...8.1461T.
- ↑ "Digestive chitinolytic activity in marine fishes of Monterey Bay, California". Comparative Biochemistry and Physiology. Part A, Molecular & Integrative Physiology 139 (3): 351–8. November 2004. doi:10.1016/j.cbpb.2004.09.020. PMID 15556391.
- ↑ "Human gastric juice contains chitinase that can degrade chitin". Annals of Nutrition & Metabolism 51 (3): 244–51. 2007. doi:10.1159/000104144. PMID 17587796.
- ↑ "Purification and characterization of human chitotriosidase, a novel member of the chitinase family of proteins". The Journal of Biological Chemistry 270 (5): 2198–202. February 1995. doi:10.1074/jbc.270.5.2198. PMID 7836450.
- ↑ "Chitinase activity in human serum and leukocytes". Infection and Immunity 63 (12): 4770–3. December 1995. doi:10.1128/IAI.63.12.4770-4773.1995. PMID 7591134.
- ↑ "Human cartilage gp-39, a major secretory product of articular chondrocytes and synovial cells, is a mammalian member of a chitinase protein family". The Journal of Biological Chemistry 268 (34): 25803–10. December 1993. doi:10.1016/S0021-9258(19)74461-5. PMID 8245017.
- ↑ "The chitinase 3-like protein human cartilage glycoprotein 39 (HC-gp39) stimulates proliferation of human connective-tissue cells and activates both extracellular signal-regulated kinase- and protein kinase B-mediated signalling pathways". The Biochemical Journal 365 (Pt 1): 119–26. July 2002. doi:10.1042/BJ20020075. PMID 12071845.
- ↑ "Characterization of human phagocyte-derived chitotriosidase, a component of innate immunity". International Immunology 17 (11): 1505–12. November 2005. doi:10.1093/intimm/dxh328. PMID 16214810.
- ↑ "Polymorphisms and haplotypes of acid mammalian chitinase are associated with bronchial asthma". American Journal of Respiratory and Critical Care Medicine 172 (12): 1505–9. December 2005. doi:10.1164/rccm.200506-890OC. PMID 16179638.
- ↑ "Increased lungkine and chitinase levels in allergic airway inflammation: a proteomics approach". Proteomics 5 (11): 2799–807. July 2005. doi:10.1002/pmic.200401169. PMID 15996009.
- ↑ "Chitinases and chitinase-like proteins in T(H)2 inflammation and asthma". The Journal of Allergy and Clinical Immunology 116 (3): 497–500. September 2005. doi:10.1016/j.jaci.2005.06.028. PMID 16159614.
- ↑ "Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation". Science 304 (5677): 1678–82. June 2004. doi:10.1126/science.1095336. PMID 15192232. Bibcode: 2004Sci...304.1678Z.
- ↑ "A chitinase-like protein in the lung and circulation of patients with severe asthma". The New England Journal of Medicine 357 (20): 2016–27. November 2007. doi:10.1056/NEJMoa073600. PMID 18003958.
- ↑ "Infections and allergy - helminths, hygiene and host immune regulation". Current Opinion in Immunology 17 (6): 656–61. December 2005. doi:10.1016/j.coi.2005.09.001. PMID 16202576.
- ↑ "Review article: helminths as therapeutic agents for inflammatory bowel disease". Alimentary Pharmacology & Therapeutics 19 (2): 167–77. January 2004. doi:10.1111/j.0269-2813.2004.01803.x. PMID 14723608.
- ↑ "Causality or coincidence: may the slow disappearance of helminths be responsible for the imbalances in immune control mechanisms?". Journal of Helminthology 77 (2): 147–53. June 2003. doi:10.1079/JOH2003176. PMID 12756068.
- ↑ "Food additives in clinical medicine". International Journal of Dermatology 14 (2): 112–4. March 1975. doi:10.1111/j.1365-4362.1975.tb01426.x. PMID 1123257.
- ↑ 35.0 35.1 "Fungal chitinases: function, regulation, and potential roles in plant/pathogen interactions". Current Genetics 62 (2): 243–54. May 2016. doi:10.1007/s00294-015-0530-x. PMID 26527115.
- ↑ "The Nag1 N-acetylglucosaminidase of Trichoderma atroviride is essential for chitinase induction by chitin and of major relevance to biocontrol". Current Genetics 43 (4): 289–95. July 2003. doi:10.1007/s00294-003-0399-y. PMID 12748812.
- ↑ "Chitinase is required for cell separation during growth of Saccharomyces cerevisiae". The Journal of Biological Chemistry 266 (29): 19758–67. October 1991. doi:10.1016/S0021-9258(18)55057-2. PMID 1918080.
- ↑ "Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates". Cell 107 (6): 739–50. December 2001. doi:10.1016/S0092-8674(01)00596-7. PMID 11747810.
- ↑ "RAM: a conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis". Molecular Biology of the Cell 14 (9): 3782–803. September 2003. doi:10.1091/mbc.E03-01-0018. PMID 12972564.
- ↑ 40.0 40.1 "Transgenic potato plants with enhanced resistance to the tomato moth, Lacanobia oleracea: growth room trials". Molecular Breeding (Springer Science+Business) 3 (1): 49–63. 1997. doi:10.1023/a:1009600321838. ISSN 1380-3743.
- ↑ "Latex-Fruit Syndrome and Class 2 Food Allergy". Division of Medical Devices, Japan. http://dmd.nihs.go.jp/latex/cross-e.html.
- ↑ 42.0 42.1 "Chitinases: An update". Journal of Pharmacy & Bioallied Sciences 5 (1): 21–9. January 2013. doi:10.4103/0975-7406.106559. PMID 23559820.
External links
- Chitinase at the US National Library of Medicine Medical Subject Headings (MeSH)
- The X-ray structure of a chitinase from the pathogenic fungus Coccidioides immitis
 |
