Biology:Hexosaminidase
| β-N-Acetylhexosaminidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
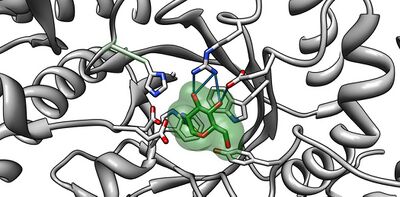
 Hexosaminidase A (Hex A) | |||||||||
| Identifiers | |||||||||
| EC number | 3.2.1.52 | ||||||||
| CAS number | 9012-33-3 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Hexosaminidase (EC 3.2.1.52, β-acetylaminodeoxyhexosidase, N-acetyl-β-D-hexosaminidase, N-acetyl-β-hexosaminidase, N-acetyl hexosaminidase, β-hexosaminidase, β-acetylhexosaminidinase, β-D-N-acetylhexosaminidase, β-N-acetyl-D-hexosaminidase, β-N-acetylglucosaminidase, hexosaminidase A, N-acetylhexosaminidase, β-D-hexosaminidase) is an enzyme involved in the hydrolysis of terminal N-acetyl-D-hexosamine residues in N-acetyl-β-D-hexosaminides.[1][2][3][4]
Elevated levels of hexosaminidase in blood and/or urine have been proposed as a biomarker of relapse in the treatment of alcoholism.[5]
Hereditary inability to form functional hexosaminidase enzymes are the cause of lipid storage disorders Tay-Sachs disease and Sandhoff disease.[6]
Isoenzymes and genes
Lysosomal A, B, and S isoenzymes
Functional lysosomal β-hexosaminidase enzymes are dimeric in structure. Three isoenzymes are produced through the combination of α and β subunits to form any one of three active dimers:[7]
| hexosaminidase isoenzyme |
subunit composition | function |
|---|---|---|
| A | α/β heterodimer | only isoenzyme that can hydrolyze GM2 ganglioside in vivo |
| B | β/β homodimer | exists in tissues but no known physiological function |
| S | α/α homodimer | exists in tissues but no known physiological function |
The α and β subunits are encoded by separate genes, HEXA and HEXB respectively. β-Hexosaminidase and the cofactor GM2 activator protein catalyze the degradation of the GM2 gangliosides and other molecules containing terminal N-acetyl hexosamines.[8] Gene mutations in HEXB often result in Sandhoff disease; whereas, mutations in HEXA decrease the hydrolysis of GM2 gangliosides, which is the main cause of Tay–Sachs disease.[9]
|
| ||||||||||||||||||||||||||||||||||||||||||||
Function
Even though the α and β subunits of lysosomal hexosaminidase can both cleave GalNAc residues, only the α subunit is able to hydrolyze GM2 gangliosides because of a key residue, Arg-424, and a loop structure that forms from the amino acid sequence in the alpha subunit. The loop in the α subunit, consisting of Gly-280, Ser-281, Glu-282, and Pro-283 which is absent in the β subunit, serves as an ideal structure for the binding of the GM2 activator protein (GM2AP), and arginine is essential for binding the N-acetyl-neuraminic acid residue of GM2 gangliosides. The GM2 activator protein transports GM2 gangliosides and presents the lipids to hexosaminidase, so a functional hexosaminidase enzyme is able to hydrolyze GM2 gangliosides into GM3 gangliosides by removing the N-acetylgalactosamine (GalNAc) residue from GM2 gangliosides.[10]
Mechanism of action
A Michaelis complex consisting of a glutamate residue, a GalNAc residue on the GM2 ganglioside, and an aspartate residue leads to the formation of an oxazolinium ion intermediate. A glutamate residue (α Glu-323/β Glu-355) works as an acid by donating its hydrogen to the glycosidic oxygen atom on the GalNAc residue. An aspartate residue (α Asp-322/β Asp-354) positions the C2-acetamindo group so that it can be attacked by the nucleophile (N-acetamido oxygen atom on carbon 1 of the substrate). The aspartate residue stabilizes the positive charge on the nitrogen atom in the oxazolinium ion intermediate. Following the formation of the oxazolinium ion intermediate, water attacks the electrophillic acetal carbon. Glutamate acts as a base by deprotonating the water leading to the formation of the product complex and the GM3ganglioside.[10]
Gene mutations resulting in Tay–Sachs disease
There are numerous mutations that lead to hexosaminidase deficiency including gene deletions, nonsense mutations, and missense mutations. Tay–Sachs disease occurs when hexosaminidase A loses its ability to function. People with Tay–Sachs disease are unable to remove the GalNAc residue from the GM2 ganglioside, and as a result, they end up storing 100 to 1000 times more GM2 gangliosides in the brain than the unaffected person. Over 100 different mutations have been discovered just in infantile cases of Tay–Sachs disease alone.[11]
The most common mutation, which occurs in over 80 percent of Tay–Sachs patients, results from a four base pair addition (TATC) in exon 11 of the Hex A gene. This insertion leads to an early stop codon, which causes the Hex A deficiency.[12]
Children born with Tay–Sachs usually die between two and four years of age from aspiration and pneumonia. Tay–Sachs causes cerebral degeneration and blindness. Patients also experience flaccid extremities and seizures. At present there has been no cure or effective treatment of Tay–Sachs disease.[11]
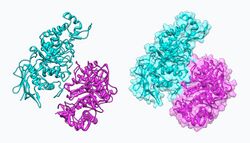
NAG-thiazoline, NGT, acts as mechanism based inhibitor of hexosaminidase A. In patients with Tay–Sachs disease (misfolded hexosaminidase A), NGT acts as a molecular chaperone by binding in the active site of hexosaminidase A which helps create a properly folded hexosaminidase A. The stable dimer conformation of hexosaminidase A has the ability to leave the endoplasmic reticulum and is directed to the lysosome where it can perform the degradation of GM2 gangliosides.[10] The two subunits of hexosaminidase A are shown below:
Cytosolic C and D isozymes
The bifunctional protein NCOAT (nuclear cytoplasmic O-GlcNAcase and acetyltransferase) that is encoded by the MGEA5 gene possesses both hexosaminidase and histone acetyltransferase activities.[13] NCOAT is also known as hexosaminidase C[14] and has distinct substrate specificities compared to lysosomal hexosaminidase A.[15] A single-nucleotide polymorphism in the human O-GlcNAcase gene is linked to diabetes mellitus type 2.[16]
A fourth mammalian hexosaminidase polypeptide which has been designated hexosaminidase D (HEXDC) has recently been identified.[17]
|
| ||||||||||||||||||||||||||||||||||||||||||||
References
- ↑ Cabezas JA (August 1989). "Some comments on the type references of the official nomenclature (IUB) for β-N-acetylglucosaminidase, β-N-acetylhexosaminidase and β-N-acetylgalactosaminidase". Biochem. J. 261 (3): 1059–60. doi:10.1042/bj2611059b. PMID 2529847.
- ↑ "Purification and properties of β-N-acetylhexosaminidase from the mollusc Helicella ericetorum Müller". Biochem. J. 175 (2): 743–50. November 1978. doi:10.1042/bj1750743. PMID 33660.
- ↑ "Isolation of β-N-acetylhexosaminidase, β-N-acetylglucosaminidase, and β-N-acetylgalactosaminidase from calf brain". Biochemistry 6 (9): 2775–82. September 1967. doi:10.1021/bi00861a018. PMID 6055190.
- ↑ "Studies on the glycosidases of jack bean meal. 3. Crystallization and properties of β-N-acetylhexosaminidase". J. Biol. Chem. 245 (19): 5153–60. October 1970. doi:10.1016/S0021-9258(18)62830-3. PMID 5506280.
- ↑ "Biomarkers of Heavy Drinking". August 2004. https://pubs.niaaa.nih.gov/publications/AssessingAlcohol/biomarkers.htm.
- ↑ Demczko, Matt. "Tay-Sachs Disease and Sandhoff Disease". https://www.msdmanuals.com/en-in/professional/pediatrics/inherited-disorders-of-metabolism/tay-sachs-disease-and-sandhoff-disease.
- ↑ "Direct determination of the substrate specificity of the alpha-active site in heterodimeric beta-hexosaminidase". Biochemistry 35 (13): 3963–9. April 1996. doi:10.1021/bi9524575. PMID 8672428.
- ↑ "NAG-thiazoline, an N-acetylbeta-hexosaminidase inhibitor that implicates acetamido participation". J. Am. Chem. Soc. 118 (28): 6804–6805. 1996. doi:10.1021/ja960826u.
- ↑ "Crystal Structure of Human β-Hexosaminidase B: Understanding the Molecular Basis of Sandhoff and Tay–Sachs Disease". J. Mol. Biol. 327 (5): 1093–109. April 2003. doi:10.1016/S0022-2836(03)00216-X. PMID 12662933.
- ↑ 10.0 10.1 10.2 10.3 10.4 10.5 10.6 "Crystallographic Structure of Human β-Hexosaminidase A: Interpretation of Tay-Sachs Mutations and Loss of GM2 Ganglioside Hydrolysis". J. Mol. Biol. 359 (4): 913–29. June 2006. doi:10.1016/j.jmb.2006.04.004. PMID 16698036.
- ↑ 11.0 11.1 "Part Thirteen Lipid Storage Disorders: Tay-Sachs disease/hexosaminidase A deficiency". Atlas of metabolic diseases. London: Hodder Arnold. 2005. pp. 539–546. ISBN 0-340-80970-1.
- ↑ "The molecular basis of HEXA mRNA deficiency caused by the most common Tay–Sachs disease mutation". Am. J. Hum. Genet. 56 (3): 716–24. March 1995. PMID 7887427.
- ↑ "The histone acetyltransferase NCOAT contains a zinc finger-like motif involved in substrate recognition". J. Biol. Chem. 281 (7): 3918–25. February 2006. doi:10.1074/jbc.M510485200. PMID 16356930.
- ↑ "Studies on human N-acetyl-Beta-d-hexosaminidase C separated from neonatal brain". Biochem. J. 155 (1): 205–8. April 1976. doi:10.1042/bj1550205. PMID 945735.
- ↑ "Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain". J. Biol. Chem. 276 (13): 9838–45. March 2001. doi:10.1074/jbc.M010420200. PMID 11148210.
- ↑ "Caenorhabditis elegans ortholog of a diabetes susceptibility locus: oga-1 (O-GlcNAcase) knockout impacts O-GlcNAc cycling, metabolism, and dauer". Proc. Natl. Acad. Sci. U.S.A. 103 (32): 11952–7. August 2006. doi:10.1073/pnas.0601931103. PMID 16882729. Bibcode: 2006PNAS..10311952F.
- ↑ "Mammalian cells contain a second nucleocytoplasmic hexosaminidase". Biochem. J. 419 (1): 83–90. April 2009. doi:10.1042/BJ20081630. PMID 19040401.
External links
- GeneReviews/NCBI/NIH/UW entry on hexosaminidase A deficiency, Tay–Sachs disease
- hexosaminidase A at the US National Library of Medicine Medical Subject Headings (MeSH)
- EC 3.2.1.52
 |