Biology:Inoviridae
| Inoviridae | |
|---|---|
| |
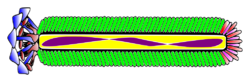
| Representation of the filamentous phage M13. Blue: Coat Protein pIII Brown: Coat Proteín pVI Red: Coat Protein pVII Limegreen: Coat Protein pVIII Fuchsia: Coat Proteín pIX Purple: Single Stranded DNA | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Monodnaviria |
| Kingdom: | Loebvirae |
| Phylum: | Hofneiviricota |
| Class: | Faserviricetes |
| Order: | Tubulavirales |
| Family: | Inoviridae |
| Genera | |
|
See text | |
Inoviridae is a family of viruses that infect bacteria. Members of the family are commonly called filamentous bacteriophages due to their filamentous shape that resembles a worm-like chain (long, thin and flexible, reminiscent of a length of cooked spaghetti) about 6 nm in diameter and about 1000-2000 nm long.[1][2][3][4] Filamentous bacteriophages are among the simplest living organisms known, with far fewer genes than the classical bacteriophages studied by the phage group. Its simplicity makes it an attractive model system to study fundamental aspects of molecular biology, and it has also proven useful as a tool in immunology and nanotechnology. The family contains 29 defined species, divided between 23 genera.[5][6] However, mining of genomic and metagenomic datasets using machine learning approach led to the discovery of 10,295 inovirus-like sequences in nearly all bacterial phyla across virtually every ecosystem, indicating that this group of viruses is much more diverse and widespread than originally appreciated.[7]
Characteristics
File:Inovirus (filamentous bacteriophage) assembled major coat protein, exploded view.tif The molecular structure of Ff filamentous phage was determined using a number of physical techniques, especially X-ray fiber diffraction.[2][8] Filamentous phage structures were further refined using solid-state NMR and cryo-electron microscopy.[2][9] The single-stranded Ff phage DNA runs down the central core of the phage, and is protected by a cylindrical protein coat built from thousands of identical α-helical protein subunits coded by phage gene 8. The gene 8 protein is inserted in the plasma membrane as an early step in phage assembly.[2] Some strains of phage have a "leader sequence" to promote membrane insertion, but others do not seem to need the leader sequence. The two ends of the phage are capped by a few copies of proteins that are important for infection of the host bacteria, and also for assembly of nascent phage particles. These proteins are the products of phage genes 3 and 6 at one end of the phage, and phage genes 7 and 9 at the other end. The fiber diffraction studies identified two structural classes of phage, differing in the details of the arrangement of the gene 8 protein. Class I, including strains fd, f1, M13, If1 and IKe, has a rotation axis relating the gene 8 coat proteins, whereas Class II, including strains Pf1, Pf3, Pf4 and PH75, this rotation axis is replaced by a helix axis. This technical difference has little noticeable effect on the overall phage structure, but the extent of independent diffraction data is greater for symmetry Class II than for Class I. This assisted the determination of the Class II phage Pf1 structure, and by extension the Class I structure.[8]
The DNA isolated from fd phage is single-stranded, and topologically a circle. That is, the DNA single strand extends from one end of the phage particle to the other and then back again to close the circle, although the two strands are not base-paired. This topology was assumed to extend to all other filamentous phages, but it is not the case for phage Pf4, for which the DNA in the phage is topologically linear, not circular.[9] During fd phage assembly, the phage DNA is first packaged in a linear intracellular nucleoprotein complex with many copies of the phage gene 5 replication/assembly protein, which is displaced by the gene 8 coat protein as the nascent phage is extruded across the bacterial plasma membrane without killing the bacterial host.[10][11][2][12] This assembly mechanism makes this phage a valuable system with which to study transmembrane proteins.[2][4]
Life cycle
Viral replication is cytoplasmic. Entry into the host cell is achieved by pilus-mediated adsorption into the host cell. Replication follows the ssDNA rolling circle model. DNA-templated transcription is the method of transcription. The virus exits the host cell by viral extrusion.[5]
Taxonomy
The following genera are recognized:[6]
- Affertcholeramvirus
- Bifilivirus
- Capistrivirus
- Coriovirus
- Fibrovirus
- Fibrovirus
- Habenivirus
- Habenivirus
- Habenivirus
- Habenivirus
- Infulavirus
- Inovirus
- Lineavirus
- Lineavirus
- Parhipatevirus
- Primolicivirus
- Psecadovirus
- Restivirus
- Saetivirus
- Saetivirus
- Scuticavirus
- Staminivirus
- Subteminivirus
- Tertilicivirus
- Thomixvirus
- Versovirus
- Vicialiavirus
- Villovirus
- Xylivirus
Phylogenetic trees and clades have been increasingly used to study taxonomy[13] of Inoviridae.[1][3][7][14]
Notable members
- Ff phages – these infect Escherichia coli carrying the F episome.
- Pf1 phage — bacteriophage that infects Pseudomonas aeruginosa
History
The filamentous particle seen in electron micrographs was initially interpreted as contaminating bacterial pili, but ultrasonic degradation, which breaks flexible filaments roughly in half,[15] inactivated infectivity as predicted for a filamentous phage morphology.[16] Three filamentous bacteriophages, fd, f1 and M13, were isolated and characterized by three different research groups in the early 1960s. Since these three phages differ by less than 2 percent in their DNA sequences, corresponding to changes in only a few dozen codons in the whole genome, for many purposes they can be considered to be identical.[17] Further independent characterization over the subsequent half-century was shaped by the interests of these research groups and their followers.[2]
Filamentous phages, unlike most other phages, are continually extruded through the bacterial membrane without killing the host.[12] Genetic studies on M13 using conditional lethal mutants, initiated by David Pratt and colleagues, led to description of phage gene functions.[18][19] Notably, the protein product of gene 5, which is required for synthesis of progeny single-stranded DNA, is made in large amounts in the infected bacteria,[20][21][22] and it binds to the nascent DNA to form a linear intracellular complex.[10] (The simple numbering of genes using Arabic numerals 1,2,3,4… introduced by the Pratt group is sometimes displaced by the practice, introduced by some f1 researchers, of using Roman numerals I, II, III, IV… but the gene numbers defined by the two systems are the same).
Longer (or shorter) DNA can be included in fd phage, since more (or fewer) protein subunits can be added during assembly as required to protect the DNA, making the phage convenient for genetic studies.[23] The length of the phage is also affected by the positive charge per length on the inside surface of the phage capsid.[24] M13 is widely used for research involving phage mutants, and is sometimes called the type species. The genome of fd was one of the first complete genomes to be sequenced.[25]
The taxonomy of filamentous bacteriophage was defined by Andre Lwoff and Paul Tournier as family Inophagoviridae, genus I. inophagovirus, species Inophagovirus bacterii (Inos=fiber or filament in Greek), with phage fd (Hoffmann-Berling) as the type species,[26] although this definition of fd as type species was modified in the early 1980s.[27] "Phagovirus" is tautological, and the name of the family was altered to Inoviridae and the type genus to Inovirus. This nomenclature persisted for many decades, but the number of known filamentous bacteriophages has multiplied many-fold by using a machine-learning approach, and it has been suggested that “the former Inoviridae family should be reclassified as an order, provisionally divided into 6 candidate families and 212 candidate subfamilies”.[7] Phages fd, f1, M13 and other related phages are often referred to as members of the Ff group of phages, for F specific (they infect Escherichia coli carrying the F-episome) filamentous phages, using the concept of vernacular name.[28]
Filamentous bacteriophage engineered to display immunogenic peptides are useful in immunology.[29][30][31] George Smith and Greg Winter used f1 and fd for their work on phage display for which they were awarded a share of the 2018 Nobel Prize in Chemistry. The creation and exploitation of many derivatives of M13 for a wide range of purposes, especially in materials science, has been employed by Angela Belcher and colleagues.[32] Filamentous bacteriophage can promote antibiotic tolerance by forming liquid crystalline domains[33] around bacterial cells.[34][9]
References
- ↑ 1.0 1.1 "Filamentous phages: masters of a microbial sharing economy". EMBO Reports 20 (6): e47427. June 2019. doi:10.15252/embr.201847427. PMID 30952693.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 "Filamentous Bacteriophage Proteins and Assembly". Sub-Cellular Biochemistry (Springer Singapore) 88: 261–279. 2018. doi:10.1007/978-981-10-8456-0_12. ISBN 978-981-10-8455-3. PMID 29900501.
- ↑ 3.0 3.1 "'Big things in small packages: the genetics of filamentous phage and effects on fitness of their host'". FEMS Microbiology Reviews 39 (4): 465–87. July 2015. doi:10.1093/femsre/fuu007. PMID 25670735.
- ↑ 4.0 4.1 "Filamentous Phage: Structure and Biology". Advances in Experimental Medicine and Biology (Springer International Publishing) 1053: 1–20. 2017. doi:10.1007/978-3-319-72077-7_1. ISBN 978-3-319-72076-0. PMID 29549632.
- ↑ 5.0 5.1 "Viral Zone". ExPASy. http://viralzone.expasy.org/all_by_species/113.html.
- ↑ 6.0 6.1 ICTV. "Virus Taxonomy: 2019 Release". https://talk.ictvonline.org/taxonomy/.
- ↑ 7.0 7.1 7.2 "Cryptic inoviruses revealed as pervasive in bacteria and archaea across Earth's biomes". Nature Microbiology 4 (11): 1895–1906. November 2019. doi:10.1038/s41564-019-0510-x. PMID 31332386.
- ↑ 8.0 8.1 "Molecular models and structural comparisons of native and mutant class I filamentous bacteriophages Ff (fd, f1, M13), If1 and IKe". Journal of Molecular Biology 235 (1): 260–86. January 1994. doi:10.1016/s0022-2836(05)80032-4. PMID 8289247.
- ↑ 9.0 9.1 9.2 "Phage liquid crystalline droplets form occlusive sheaths that encapsulate and protect infectious rod-shaped bacteria". Proceedings of the National Academy of Sciences of the United States of America 117 (9): 4724–4731. March 2020. doi:10.1073/pnas.1917726117. PMID 32071243.
- ↑ 10.0 10.1 "Complex of bacteriophage M13 single-stranded DNA and gene 5 protein". Journal of Molecular Biology 82 (4): 425–39. February 1974. doi:10.1016/0022-2836(74)90239-3. PMID 4594145.
- ↑ "Three-dimensional structure of complexes of single-stranded DNA-binding proteins with DNA. IKe and fd gene 5 proteins form left-handed helices with single-stranded DNA". Journal of Molecular Biology 208 (1): 57–64. July 1989. doi:10.1016/0022-2836(89)90087-9. PMID 2671388.
- ↑ 12.0 12.1 "Release of male-specific bacteriophages from surviving host bacteria". Virology 22 (3): 305–13. March 1964. doi:10.1016/0042-6822(64)90021-2. PMID 14127828.
- ↑ "The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks". Nature Microbiology 5 (5): 668–674. May 2020. doi:10.1038/s41564-020-0709-x. PMID 32341570.
- ↑ "Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids". Nature Communications 10 (1): 3425. July 2019. doi:10.1038/s41467-019-11433-0. PMID 31366885.
- ↑ "Studies on the sonic degradation of deoxyribonucleic acid". Biophysical Journal 2 (3): 235–47. May 1962. doi:10.1016/S0006-3495(62)86852-0. PMID 13894963. Bibcode: 1962BpJ.....2..235F.
- ↑ "Physical and Chemical Properties of Two New Small Bacteriophages". Nature 197 (4866): 517–518. 1963. doi:10.1038/197517b0. Bibcode: 1963Natur.197..517M.
- ↑ "Similarities and differences within members of the Ff family of filamentous bacteriophage viruses". The Journal of Physical Chemistry. B 115 (51): 15370–9. December 2011. doi:10.1021/jp2079742. PMID 22085310. https://pubs.acs.org/doi/10.1021/jp2079742.
- ↑ "Conditional lethal mutants of the small filamentous coliphage M13. I. Isolation, complementation, cell killing, time of cistron action". Virology 30 (3): 397–410. November 1966. doi:10.1016/0042-6822(66)90118-8. PMID 5921643.
- ↑ "Conditional lethal mutants of the small filamentous coliphage M13. II. Two genes for coat proteins". Virology 39 (1): 42–53. September 1969. doi:10.1016/0042-6822(69)90346-8. PMID 5807970.
- ↑ "Genetic control of bacteriophage M13 DNA synthesis". Journal of Molecular Biology 37 (1): 181–200. October 1968. doi:10.1016/0022-2836(68)90082-X. PMID 4939035.
- ↑ "The proteins of bacteriophage M13". Proceedings of the National Academy of Sciences of the United States of America 62 (3): 800–7. March 1969. doi:10.1073/pnas.62.3.800. PMID 5257006.
- ↑ "Isolation and characterization of gene 5 protein of filamentous bacterial viruses". Journal of Molecular Biology 68 (1): 139–52. July 1972. doi:10.1016/0022-2836(72)90269-0. PMID 4115107.
- ↑ "Transposition of a DNA sequence determining kanamycin resistance into the single-stranded genome of bacteriophage fd". Molecular & General Genetics 159 (2): 171–8. February 1978. doi:10.1007/bf00270890. PMID 345091.
- ↑ "Regulation of filamentous bacteriophage length by modification of electrostatic interactions between coat protein and DNA". Journal of Molecular Biology 217 (2): 223–7. January 1991. doi:10.1016/0022-2836(91)90534-d. PMID 1992159.
- ↑ "Nucleotide sequence of bacteriophage fd DNA". Nucleic Acids Research 5 (12): 4495–503. December 1978. doi:10.1093/nar/5.12.4495. PMID 745987.
- ↑ "The classification of viruses". Annual Review of Microbiology 20 (1): 45–74. 1966. doi:10.1146/annurev.mi.20.100166.000401. PMID 5330240.
- ↑ "Classification and nomenclature of viruses. Fourth report of the International Committee on Taxonomy of Viruses". Intervirology 17 (1-3): 1–199. 1982. doi:10.1159/000149278. PMID 6811498.
- ↑ "What's in a virus name?". Nature 209 (5022): 450–4. January 1966. doi:10.1038/209450a0. PMID 5919575. Bibcode: 1966Natur.209..450G.
- ↑ "Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface". Science 228 (4705): 1315–7. June 1985. doi:10.1126/science.4001944. PMID 4001944. https://www.sciencemag.org/lookup/doi/10.1126/science.4001944.
- ↑ "Filamentous bacteriophage fd as an antigen delivery system in vaccination". International Journal of Molecular Sciences 13 (4): 5179–94. 2012-04-24. doi:10.3390/ijms13045179. PMID 22606037. http://dx.doi.org/10.3390/ijms13045179.
- ↑ "Phage Display Libraries: From Binders to Targeted Drug Delivery and Human Therapeutics". Molecular Biotechnology 61 (4): 286–303. April 2019. doi:10.1007/s12033-019-00156-8. PMID 30729435.
- ↑ "Constructing Multifunctional Virus-Templated Nanoporous Composites for Thin Film Solar Cells: Contributions of Morphology and Optics to Photocurrent Generation". The Journal of Physical Chemistry C 119 (25): 13987–4000. June 2015. doi:10.1021/acs.jpcc.5b00295. ISSN 1932-7447.
- ↑ "Filamentous Phages As a Model System in Soft Matter Physics". Frontiers in Microbiology 7: 1013. 2016-06-30. doi:10.3389/fmicb.2016.01013. PMID 27446051.
- ↑ "Pseudomonas aeruginosa biofilm matrix into a liquid crystal". Microbial Cell 3 (1): 49–52. December 2015. doi:10.15698/mic2016.01.475. PMID 28357315.
External links
Wikidata ☰ Q3502731 entry

