Biology:Origin recognition complex
| Origin recognition complex subunit 2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | ORC2 | ||||||||
| Pfam | PF04084 | ||||||||
| InterPro | IPR007220 | ||||||||
| |||||||||
| Origin recognition complex (ORC) subunit 3 N-terminus | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | ORC3_N | ||||||||
| Pfam | PF07034 | ||||||||
| InterPro | IPR010748 | ||||||||
| |||||||||
| Origin recognition complex subunit 6 (ORC6) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | ORC6 | ||||||||
| Pfam | PF05460 | ||||||||
| InterPro | IPR008721 | ||||||||
| |||||||||
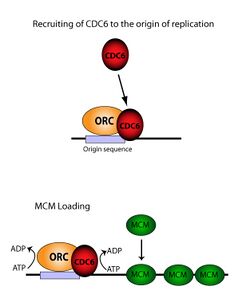
In molecular biology, origin recognition complex (ORC) is a multi-subunit DNA binding complex (6 subunits) that binds in all eukaryotes and archaea in an ATP-dependent manner to origins of replication. The subunits of this complex are encoded by the ORC1, ORC2, ORC3, ORC4, ORC5 and ORC6 genes.[1][2][3] ORC is a central component for eukaryotic DNA replication, and remains bound to chromatin at replication origins throughout the cell cycle.[4]
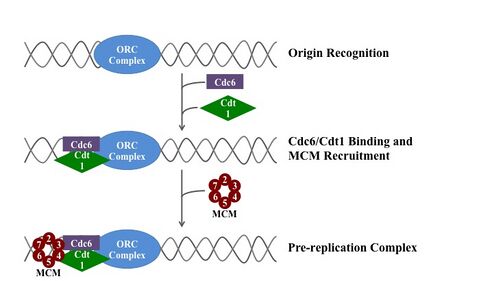
ORC directs DNA replication throughout the genome and is required for its initiation.[5][6][7] ORC and Noc3p bound at replication origins serve as the foundation for assembly of the pre-replication complex (pre-RC), which includes Cdc6, Tah11 (a.k.a. Cdt1), and the Mcm2-Mcm7 complex.[8][9][10][11] Pre-RC assembly during G1 is required for replication licensing of chromosomes prior to DNA synthesis during S phase.[12][13][14] Cell cycle-regulated phosphorylation of Orc2, Orc6, Cdc6, and MCM by the cyclin-dependent protein kinase Cdc28 regulates initiation of DNA replication, including blocking reinitiation in G2/M phase.[4][15][16][17]
The ORC is present throughout the cell cycle bound to replication origins, but is only active in late mitosis and early G1.
In yeast, ORC also plays a role in the establishment of silencing at the mating-type loci Hidden MAT Left (HML) and Hidden MAT Right (HMR).[5][6][7] ORC participates in the assembly of transcriptionally silent chromatin at HML and HMR by recruiting the Sir1 silencing protein to the HML and HMR silencers.[7][18][19]
Both Orc1 and Orc5 bind ATP, though only Orc1 has ATPase activity.[20] The binding of ATP by Orc1 is required for ORC binding to DNA and is essential for cell viability.[11] The ATPase activity of Orc1 is involved in formation of the pre-RC.[21][22][23] ATP binding by Orc5 is crucial for the stability of ORC as a whole. Only the Orc1-5 subunits are required for origin binding; Orc6 is essential for maintenance of pre-RCs once formed.[24] Interactions within ORC suggest that Orc2-3-6 may form a core complex.[4] A 2020 report suggests that budding yeast ORC dimerizes in a cell cycle dependent manner to control licensing.[25][26]
Proteins
File:Electrochemical-Regulation-of-Budding-Yeast-Polarity-pbio.1002029.s012.ogv The following proteins are present in the ORC:
| S. cerevisiae | S. pombe | D. melanogaster | Vertebrates |
|---|---|---|---|
| ORC 1-6 | ORC 1-6 | ORC 1-6 | ORC 1-6 |
| Cdc6 | Cdc18 | Cdc6 | Cdc6 |
| Cdt1/Tah11/Sid2 | Cdt1 | DUP | Cdt1/RLF-B |
| Mcm2 | Mcm2/Cdc19/Nda1 | Mcm2 | Mcm2 |
| Mcm3 | Mcm3 | Mcm3 | Mcm3 |
| Cdc54/Mcm4 | Cdc21 | DPA | Mcm4 |
| Cdc46/Mcm5 | Mcm5/Nda4 | Mcm5 | Mcm5 |
| Mcm6 | Mcm6/Mis5 | Mcm6 | Mcm6 |
| Cdc47/Mcm7 | Mcm7 | Mcm7 | mcm7 |
Archaea feature a simplified version of the ORC, Mcm, and as a consequence the combined pre-RC. Instead of using six different mcm proteins to form a pseudo-symmetrical heterohexamer, all six subunits in the archaeal MCM are the same. They usually have multiple proteins that are homologous to both Cdc6 and Orc1, some of which perform the function of both. Unlike eukaryotic Orc, they do not always form a complex. In fact, they have divergent complex structures when these do form. Sulfolobus islandicus also uses a Cdt1 homologue to recognize one of its replication origins.[28]
Autonomously replicating sequences
Budding yeast
Autonomously Replicating Sequences (ARS), first discovered in budding yeast, are integral to the success of the ORC. These 100-200bp sequences facilitate replication activity during S phase. ARSs can be placed at any novel location of the chromosomes of budding yeast and will facilitate replication from those sites. A highly conserved sequence of 11bp (known as the A element) is thought to be essential for origin function in budding yeast.[27] The ORC was originally identified by its ability to bind to the A element of the ARS in budding yeast.
Animals
Animal cells contain a much more cryptic version of an ARS, with no conserved sequences found as of yet. Here, replication origins gather into bundles called replicon clusters. Each cluster's replicons are similar in length, but individual clusters have replicons of varying length. These replicons all have similar basic residues to which the ORC binds, which in many ways mimic the conserved 11bp A element. All of these clusters are simultaneously activated during S phase.[27]
Role in pre-RC assembly
The ORC is essential for the loading of MCM complexes (Pre-RC) onto DNA. This process is dependent on the ORC, Noc3, Cdc6, and Cdt1 – involving several ATP controlled recruiting events. First, the ORC, Noc3p and Cdc6 form a complex on origin DNA (marked by ARS type regions). New ORC/Noc3/Cdc6 complexes then recruit Cdt1/Mcm2-7 molecules to the site. Once this massive ORC/Noc3/Cdc6/Cdt1/Mcm2-7 complex is formed, the ORC/Noc3/Cdc6/Cdt1 molecules work together to load Mcm2-7 onto the DNA itself by hydrolysis of ATP by Cdc6. Cdc6's phosphorylative activity is dependent on both the ORC and origin DNA. This leads to Cdt1 having decreased stability on the DNA and falling off of the complex leading to Mcm2-7 loading on to the DNA.[29][27][30][31] The structure of the ORC, MCM, as well as the intermediate OCCM complex has been resolved.[32]
Origin binding activity
Although the ORC is composed of six discrete subunits, only one of these has been found to be significant - ORC1. In vivo studies have shown that Lys-263 and Arg-367 are the basic residues responsible for faithful ORC loading. These molecules represent the above-mentioned ARS.[33] ORC1 interacts with ATP and these basic residues in order to bind the ORC to origin DNA. It has been established that this occurs far before replication, and that the ORC itself is already bound to Origin DNA by the time any Mcm2-7 loading occurs.[31] When Mcm2-7 is first loaded it completely encircles the DNA and helicase activity is inhibited. In S phase, the Mcm2-7 complex interacts with helicase cofactors Cdc45 and GINS to isolate a single DNA strand, unwind the origin, and begin replication down the chromosome. In order to have bidirectional replication, this process happens twice at an origin. Both loading events are mediated by one ORC via an identical process as the first.[34]
See also
- Cyclin dependant kinases (CDK)
- Cyclins
- DNA helicase
- DnaA
- Pre-replication complex
References
- ↑ Origin+Recognition+Complex at the US National Library of Medicine Medical Subject Headings (MeSH)
- ↑ "Initiation of DNA replication in eukaryotic cells". Annual Review of Cell and Developmental Biology 13: 293–332. 1997. doi:10.1146/annurev.cellbio.13.1.293. PMID 9442876.
- ↑ Multiple functions of the origin recognition complex. International Review of Cytology. 256. 2007. pp. 69–109. doi:10.1016/S0074-7696(07)56003-1. ISBN 9780123737007.
- ↑ 4.0 4.1 4.2 "Yeast two-hybrid analysis of the origin recognition complex of Saccharomyces cerevisiae: interaction between subunits and identification of binding proteins". FEMS Yeast Research 7 (8): 1263–9. December 2007. doi:10.1111/j.1567-1364.2007.00298.x. PMID 17825065.
- ↑ 5.0 5.1 "ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex". Nature 357 (6374): 128–34. May 1992. doi:10.1038/357128a0. PMID 1579162. Bibcode: 1992Natur.357..128B.
- ↑ 6.0 6.1 "The multidomain structure of Orc1p reveals similarity to regulators of DNA replication and transcriptional silencing". Cell 83 (4): 563–8. November 1995. doi:10.1016/0092-8674(95)90096-9. PMID 7585959.
- ↑ 7.0 7.1 7.2 "Cell cycle execution point analysis of ORC function and characterization of the checkpoint response to ORC inactivation in Saccharomyces cerevisiae". Genes to Cells 11 (6): 557–73. June 2006. doi:10.1111/j.1365-2443.2006.00967.x. PMID 16716188.
- ↑ "Noc3p, a bHLH Protein, Plays an Integral Role in the Initiation of DNA Replication in Budding Yeast". Cell 109 (7): 849–860. June 2002. doi:10.1016/s0092-8674(02)00805-x. PMID 12110182.
- ↑ "The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators". Proceedings of the National Academy of Sciences of the United States of America 92 (6): 2224–8. March 1995. doi:10.1073/pnas.92.6.2224. PMID 7892251. Bibcode: 1995PNAS...92.2224R.
- ↑ "Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC". The EMBO Journal 14 (11): 2631–41. June 1995. doi:10.1002/j.1460-2075.1995.tb07261.x. PMID 7781615.
- ↑ 11.0 11.1 "ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA". Nature Structural & Molecular Biology 12 (11): 965–71. November 2005. doi:10.1038/nsmb1002. PMID 16228006.
- ↑ "Regulation of chromosome replication". Annual Review of Biochemistry 69: 829–80. 2000. doi:10.1146/annurev.biochem.69.1.829. PMID 10966477.
- ↑ "DNA replication in eukaryotic cells". Annual Review of Biochemistry 71: 333–74. 2002. doi:10.1146/annurev.biochem.71.110601.135425. PMID 12045100.
- ↑ "Origin recognition and the chromosome cycle". FEBS Letters 579 (4): 877–84. February 2005. doi:10.1016/j.febslet.2004.12.011. PMID 15680967.
- ↑ "Binding of cyclin-dependent kinases to ORC and Cdc6p regulates the chromosome replication cycle". Proceedings of the National Academy of Sciences of the United States of America 98 (20): 11211–7. September 2001. doi:10.1073/pnas.201387198. PMID 11572976.
- ↑ "Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms". Nature 411 (6841): 1068–73. June 2001. doi:10.1038/35082600. PMID 11429609. Bibcode: 2001Natur.411.1068N.
- ↑ "Disruption of mechanisms that prevent rereplication triggers a DNA damage response". Molecular and Cellular Biology 25 (15): 6707–21. August 2005. doi:10.1128/MCB.25.15.6707-6721.2005. PMID 16024805.
- ↑ "Role of interactions between the origin recognition complex and SIR1 in transcriptional silencing". Nature 381 (6579): 251–3. May 1996. doi:10.1038/381251a0. PMID 8622770. Bibcode: 1996Natur.381..251T.
- ↑ "The origin recognition complex, SIR1, and the S phase requirement for silencing". Science 276 (5318): 1547–51. June 1997. doi:10.1126/science.276.5318.1547. PMID 9171055.
- ↑ "Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex". Cell 88 (4): 493–502. February 1997. doi:10.1016/S0092-8674(00)81889-9. PMID 9038340.
- ↑ "ATP bound to the origin recognition complex is important for preRC formation". Proceedings of the National Academy of Sciences of the United States of America 98 (15): 8361–7. July 2001. doi:10.1073/pnas.131006898. PMID 11459976. Bibcode: 2001PNAS...98.8361K.
- ↑ "ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication". Molecular Cell 16 (6): 967–78. December 2004. doi:10.1016/j.molcel.2004.11.038. PMID 15610739.
- ↑ "Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase". Molecular Cell 21 (1): 29–39. January 2006. doi:10.1016/j.molcel.2005.11.023. PMID 16387651.
- ↑ "An essential role for Orc6 in DNA replication through maintenance of pre-replicative complexes". The EMBO Journal 25 (21): 5150–8. November 2006. doi:10.1038/sj.emboj.7601391. PMID 17053779.
- ↑ "Replication initiation: Implications in genome integrity". DNA Repair 103: 103131. July 2021. doi:10.1016/j.dnarep.2021.103131. PMID 33992866.
- ↑ "An Essential and Cell-Cycle-Dependent ORC Dimerization Cycle Regulates Eukaryotic Chromosomal DNA Replication". Cell Reports 30 (10): 3323–3338.e6. March 2020. doi:10.1016/j.celrep.2020.02.046. PMID 32160540.
- ↑ 27.0 27.1 27.2 27.3 Morgan, David (2007). The Cell Cycle: Principles of Control. Primers in Biology. pp. 62–75. ISBN 978-0878935086.
- ↑ "Diversity of DNA Replication in the Archaea". Genes 8 (2): 56. January 2017. doi:10.3390/genes8020056. PMID 28146124.
- ↑ "An ORC/Cdc6/MCM2-7 complex is formed in a multistep reaction to serve as a platform for MCM double-hexamer assembly". Molecular Cell 50 (4): 577–88. May 2013. doi:10.1016/j.molcel.2013.03.026. PMID 23603117.
- ↑ "Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase". Molecular Cell 21 (1): 29–39. January 2006. doi:10.1016/j.molcel.2005.11.023. PMID 16387651.
- ↑ 31.0 31.1 "ATPase-dependent cooperative binding of ORC and Cdc6 to origin DNA". Nature Structural & Molecular Biology 12 (11): 965–71. November 2005. doi:10.1038/nsmb1002. PMID 16228006.
- ↑ "Structural basis of Mcm2-7 replicative helicase loading by ORC-Cdc6 and Cdt1". Nature Structural & Molecular Biology 24 (3): 316–324. March 2017. doi:10.1038/nsmb.3372. PMID 28191893.
- ↑ "Specific binding of eukaryotic ORC to DNA replication origins depends on highly conserved basic residues". Scientific Reports 5: 14929. October 2015. doi:10.1038/srep14929. PMID 26456755. Bibcode: 2015NatSR...514929K.
- ↑ "Single-Molecule Visualization of MCM2-7 DNA Loading: Seeing Is Believing". Cell 161 (3): 429–430. April 2015. doi:10.1016/j.cell.2015.04.006. PMID 25910200.
Further reading
- "DNA replication in eukaryotic cells". Annual Review of Biochemistry (Annual Reviews) 71: 333–74. July 2002. doi:10.1146/annurev.biochem.71.110601.135425. PMID 12045100. "A comprehensive review of molecular DNA replication".
 |