Biology:Protein kinase
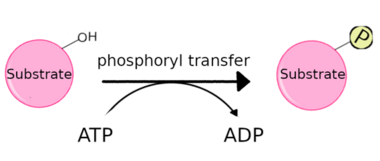
A protein kinase is a kinase which selectively modifies other proteins by covalently adding phosphates to them (phosphorylation) as opposed to kinases which modify lipids, carbohydrates, or other molecules. Phosphorylation usually results in a functional change of the target protein (substrate) by changing enzyme activity, cellular location, or association with other proteins. The human genome contains about 500 protein kinase genes and they constitute about 2% of all human genes.[1] There are two main types of protein kinase. The great majority are serine/threonine kinases, which phosphorylate the hydroxyl groups of serines and threonines in their targets. Most of the others are tyrosine kinases, although additional types exist.[2] Protein kinases are also found in bacteria and plants. Up to 30% of all human proteins may be modified by kinase activity, and kinases are known to regulate the majority of cellular pathways, especially those involved in signal transduction.
Chemical activity
The chemical activity of a protein kinase involves removing a phosphate group from ATP and covalently attaching it to one of three amino acids that have a free hydroxyl group. Most kinases act on both serine and threonine, others act on tyrosine, and a number (dual-specificity kinases) act on all three.[3] There are also protein kinases that phosphorylate other amino acids, including histidine kinases that phosphorylate histidine residues.[4]
Structure
Eukaryotic protein kinases are enzymes that belong to a very extensive family of proteins that share a conserved catalytic core.[5][6][7][8] The structures of over 280 human protein kinases have been determined.[9]
There are a number of conserved regions in the catalytic domain of protein kinases. In the N-terminal extremity of the catalytic domain there is a glycine-rich stretch of residues in the vicinity of a lysine amino acid, which has been shown to be involved in ATP binding. In the central part of the catalytic domain, there is a conserved aspartic acid, which is important for the catalytic activity of the enzyme.[10]
Serine/threonine-specific protein kinases
Serine/threonine protein kinases (EC 2.7.11.1) phosphorylate the OH group of serine or threonine (which have similar side chains). Activity of these protein kinases can be regulated by specific events (e.g., DNA damage), as well as numerous chemical signals, including cAMP/cGMP, diacylglycerol, and Ca2+/calmodulin. One very important group of protein kinases are the MAP kinases (acronym from: "mitogen-activated protein kinases"). Important subgroups are the kinases of the ERK subfamily, typically activated by mitogenic signals, and the stress-activated protein kinases JNK and p38. While MAP kinases are serine/threonine-specific, they are activated by combined phosphorylation on serine/threonine and tyrosine residues. Activity of MAP kinases is restricted by a number of protein phosphatases, which remove the phosphate groups that are added to specific serine or threonine residues of the kinase and are required to maintain the kinase in an active conformation.
Tyrosine-specific protein kinases
Tyrosine-specific protein kinases (EC 2.7.10.1 and EC 2.7.10.2) phosphorylate tyrosine amino acid residues, and like serine/threonine-specific kinases are used in signal transduction. They act primarily as growth factor receptors and in downstream signaling from growth factors.[11] Some examples include:
- Platelet-derived growth factor receptor (PDGFR)
- Epidermal growth factor receptor (EGFR)[12]
- Insulin receptor and insulin-like growth factor 1 receptor (IGF1R)
- Stem cell factor (SCF) receptor (also called c-kit, see the article on gastrointestinal stromal tumor).
Receptor tyrosine kinases
These kinases consist of extracellular domains, a transmembrane spanning alpha helix, and an intracellular tyrosine kinase domain protruding into the cytoplasm. They play important roles in regulating cell division, cellular differentiation, and morphogenesis. More than 50 receptor tyrosine kinases are known in mammals.
Structure
The extracellular domains serve as the ligand-binding part of the molecule, often inducing the domains to form homo- or heterodimers. The transmembrane element is a single α helix. The intracellular or cytoplasmic Protein kinase domain is responsible for the (highly conserved) kinase activity, as well as several regulatory functions.
Regulation
Ligand binding causes two reactions:
- Dimerization of two monomeric receptor kinases or stabilization of a loose dimer. Many ligands of receptor tyrosine kinases are multivalent. Some tyrosine receptor kinases (e.g., the platelet-derived growth factor receptor) can form heterodimers with other similar but not identical kinases of the same subfamily, allowing a highly varied response to the extracellular signal.
- Trans-autophosphorylation (phosphorylation by the other kinase in the dimer) of the kinase.
Autophosphorylation stabilizes the active conformation of the kinase domain. When several amino acids suitable for phosphorylation are present in the kinase domain (e.g., the insulin-like growth factor receptor), the activity of the kinase can increase with the number of phosphorylated amino acids; in this case, the first phosphorylation switches the kinase from "off" to "standby".
Signal transduction
The active tyrosine kinase phosphorylates specific target proteins, which are often enzymes themselves. An important target is the ras protein signal-transduction chain.
Receptor-associated tyrosine kinases
Tyrosine kinases recruited to a receptor following hormone binding are receptor-associated tyrosine kinases and are involved in a number of signaling cascades, in particular those involved in cytokine signaling (but also others, including growth hormone). One such receptor-associated tyrosine kinase is Janus kinase (JAK), many of whose effects are mediated by STAT proteins. (See JAK-STAT pathway.)
Dual-specificity protein kinases
Some kinases have dual-specificity kinase activities. For example, MEK (MAPKK), which is involved in the MAP kinase cascade, is a both a serine/threonine and tyrosine kinase.
Histidine-specific protein kinases
Histidine kinases are structurally distinct from most other protein kinases and are found mostly in prokaryotes as part of two-component signal transduction mechanisms. A phosphate group from ATP is first added to a histidine residue within the kinase, and later transferred to an aspartate residue on a 'receiver domain' on a different protein, or sometimes on the kinase itself. The aspartyl phosphate residue is then active in signaling.
Histidine kinases are found widely in prokaryotes, as well as in plants, fungi and eukaryotes. The pyruvate dehydrogenase family of kinases in animals is structurally related to histidine kinases, but instead phosphorylate serine residues, and probably do not use a phospho-histidine intermediate.
Aspartic acid/glutamic acid-specific protein kinases
Inhibitors
Deregulated kinase activity is a frequent cause of disease, in particular cancer, wherein kinases regulate many aspects that control cell growth, movement and death. Drugs that inhibit specific kinases are being developed to treat several diseases, and some are currently in clinical use, including Gleevec (imatinib) and Iressa (gefitinib).
- Anthra(1,9-cd)pyrazol-6(2H)-one
- Staurosporine
Kinase assays and profiling
Drug developments for kinase inhibitors are started from kinase assays, the lead compounds are usually profiled for specificity before moving into further tests. Many profiling services are available from fluorescent-based assays to radioisotope based detections, and competition binding assays.
References
- ↑ "The protein kinase complement of the human genome". Science 298 (5600): 1912–1934. 2002. doi:10.1126/science.1075762. PMID 12471243. Bibcode: 2002Sci...298.1912M.
- ↑ Alberts, Bruce (18 November 2014). Molecular biology of the cell (Sixth ed.). New York. pp. 819–820. ISBN 978-0-8153-4432-2. OCLC 887605755.
- ↑ "Signaling by dual specificity kinases". Oncogene 17 (11 Reviews): 1447–55. September 1998. doi:10.1038/sj.onc.1202251. PMID 9779990.
- ↑ "Mammalian protein histidine kinases". Int. J. Biochem. Cell Biol. 35 (3): 297–309. March 2003. doi:10.1016/S1357-2725(02)00257-1. PMID 12531242.
- ↑ "Genomic analysis of the eukaryotic protein kinase superfamily: a perspective". Genome Biol. 4 (5): 111. 2003. doi:10.1186/gb-2003-4-5-111. PMID 12734000.
- ↑ "Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification". FASEB J. 9 (8): 576–96. May 1995. doi:10.1096/fasebj.9.8.7768349. PMID 7768349. http://www.fasebj.org/cgi/pmidlookup?view=long&pmid=7768349.
- ↑ "Protein kinase classification". Protein Phosphorylation Part A: Protein Kinases: Assays, Purification, Antibodies, Functional Analysis, Cloning, and Expression. Methods in Enzymology. 200. 1991. 3–37. doi:10.1016/0076-6879(91)00125-G. ISBN 9780121821012.
- ↑ "Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members". Protein Phosphorylation Part A: Protein Kinases: Assays, Purification, Antibodies, Functional Analysis, Cloning, and Expression. Methods in Enzymology. 200. 1991. 38–62. doi:10.1016/0076-6879(91)00126-H. ISBN 9780121821012.
- ↑ Modi, V; Dunbrack, RL (2019-12-24). "A Structurally-Validated Multiple Sequence Alignment of 497 Human Protein Kinase Domains.". Scientific Reports 9 (1): 19790. doi:10.1038/s41598-019-56499-4. PMID 31875044. Bibcode: 2019NatSR...919790M.
- ↑ "Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase". Science 253 (5018): 407–14. July 1991. doi:10.1126/science.1862342. PMID 1862342. Bibcode: 1991Sci...253..407K.
- ↑ Higashiyama S, Iwabuki H, Morimoto C, Hieda M, Inoue H, Matsushita N. Membrane-anchored growth factors, the epidermal growth factor family: beyond receptor ligands. Cancer Sci. 2008 Feb;99(2):214-20. Review. PMID: 18271917
- ↑ Carpenter G. The EGF receptor: a nexus for trafficking and signaling. Bioessays. 2000 Aug;22(8):697-707. Review. PMID: 10918300
External links
- Human and mouse protein kinases in UniProt: classification and index
- Kinase.Com: Genomics, evolution and large-scale analysis of protein kinases (non-commercial).
- KinMutBase: A registry of disease-causing mutations in protein kinase domains
- KLIFS (Kinase-Ligand Interaction Fingerprints and Structures) Database -- analysis of kinase structures and kinase-inhibitor interactions
- KinCore: the Kinase Conformation Resource: A web resource for protein kinase sequence, structure and phylogeny
- Kinomer: A multilevel HMM library for the classification and functional annotation of eukaryotic protein kinases.
 |
