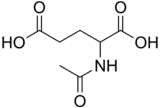
Chemistry:N-Acetylglutamic acid
| |
| Names | |
|---|---|
| IUPAC name
2-Acetamidopentanedioic acid[1]
| |
| Other names
Acetylglutamic acid[citation needed]
| |
| Identifiers | |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
|Section2=! colspan=2 style="background: #f8eaba; text-align: center;" |Properties
|-
|
| C7H11NO5
|- | Molar mass
| 189.167 g·mol−1
|- | Appearance | White crystals |-
| Density | 1 g mL−1 |- | Melting point | 191 to 194 °C (376 to 381 °F; 464 to 467 K)
|-
|
| 36 g L−1 |- |Section3=! colspan=2 style="background: #f8eaba; text-align: center;" |Hazards
|-
| colspan=2 style="text-align:left; background-color:#f1f1f1;" | Lethal dose or concentration (LD, LC):
|-
|- style="background:#f4f4f4;"
| style="padding-left:1em;" |
| >7 g kg−1 (oral, rat)
|-
|- |Section4=! colspan=2 style="background: #f8eaba; text-align: center;" |Related compounds
|-
|
|
|-
|
|
|- }} N-Acetylglutamic acid (also referred to as N-acetylglutamate, abbreviated NAG, chemical formula C7H11NO5)[2] is biosynthesized from glutamate and acetylornithine by ornithine acetyltransferase, and from glutamic acid and acetyl-CoA by the enzyme N-acetylglutamate synthase. The reverse reaction, hydrolysis of the acetyl group, is catalyzed by a specific hydrolase. It is the first intermediate involved in the biosynthesis of arginine in prokaryotes and simple eukaryotes and a regulator in the process known as the urea cycle that converts toxic ammonia to urea for excretion from the body in vertebrates.
Discovery
N-Acetylglutamic acid is an extracellular metabolite isolated from the prokaryote Rhizobium trifolii that was characterized using many structure determination techniques such as proton nuclear magnetic resonance (1H NMR) spectroscopy, Fourier-transform infrared spectroscopy, and gas chromatography-mass spectrometry.
In Rhizobium, extracellular build-up of N-acetylglutamic acid is due to metabolism involving nod factor genes on a symbiotic plasmid. When the nod factors are mutated, less N-acetylglutamic acid is produced.[3]
Biosynthesis
Prokaryotes and simple eukaryotes
In prokaryotes and simple eukaryotes, N-acetylglutamic acid can be produced by N-acetylglutamate synthase (NAGS) or ornithine acetyltransferase (OAT).
Ornithine acetyltransferase (OAT) synthesis
OAT synthesizes N-acetylglutamic acid from glutamate and acetylornithine and is the method of choice for production in prokaryotes that have the ability to synthesize the compound ornithine.[4]
N-Acetylglutamate synthase (NAGS) synthesis
N-Acetylglutamate synthase is an enzyme that serves as a replenisher of N-acetylglutamic acid to supplement any N-acetylglutamic acid lost by the cell through mitosis or degradation. NAGS synthesizes N-acetylglutamic acid by catalyzing the addition of an acetyl group from acetyl-coenzyme A to glutamate. In prokaryotes with non-cyclic ornithine production, NAGS is the sole method of N-acetylglutamic acid synthesis and is inhibited by arginine.[4] Acetylation of glutamate is thought to prevent glutamate from being used by proline biosynthesis.[5]
Vertebrates
In contrast to prokaryotes, NAGS in mammals is enhanced by arginine, along with protamines. It is inhibited by N-acetylglutamic acid and its analogues (other N-acetylated compounds).[4]
The brain also contains N-acetylglutamic acid at trace amounts, however no expression of NAGS is found. This suggests that N-acetylglutamic acid is produced by another enzyme in the brain that is yet to be determined.[4]
Biological roles
Vertebrates and mammals
In vertebrae and mammals, N-acetylglutamic acid is the allosteric activator molecule to mitochondrial carbamyl phosphate synthetase I (CPSI) which is the first enzyme in the urea cycle.[6] It triggers the production of the first urea cycle intermediate, carbamyl phosphate. CPSI is inactive when N-acetylglutamic acid is not present. In the liver and small intestines, N-acetylglutamic acid-dependent CPSI produces citrulline, the second intermediate in the urea cycle. Liver cell distribution of N-acetylglutamic acid is highest in the mitochondria at 56% of total N-acetylglutamic acid availability, 24% in the nucleus, and the remaining 20% in the cytosol. Aminoacylase I in liver and kidney cells degrades N-acetylglutamic acid to glutamate and acetate.[7] In contrast, N-acetylglutamic acid is not the allosteric cofactor to carbamyl phosphate synthetase found in the cytoplasm, which is involved in pyrimidine synthesis.[8]
N-acetylglutamic acid concentrations increase when protein consumption increases due to the accumulation of ammonia that must be secreted through the urea cycle, which supports the role of N-acetylglutamic acid as the cofactor for CPSI. Furthermore, N-acetylglutamic acid can be found in many commonly consumed foods such as soy, corn, and coffee, with cocoa powder containing a notably high concentration.[9]
Deficiency in N-acetylglutamic acid in humans is an autosomal recessive disorder that results in blockage of urea production which ultimately increases the concentration of ammonia in the blood (hyperammonemia). Deficiency can be caused by defects in the NAGS coding gene or by deficiencies in the precursors essential for synthesis.[4]
Bacteria
N-Acetylglutamic acid is the second intermediate in the arginine production pathway in Escherichia coli and is produced via NAGS.[5] In this pathway, N-acetylglutamic acid kinase (NAGK) catalyzes the phosphorylation of the gamma (third) carboxyl group of N-acetylglutamic acid using the phosphate produced by hydrolysis of adenosine triphosphate (ATP).[10]
White clover seedling roots
Rhizobium can form a symbiotic relationship with white clover seedling roots and form colonies. The extracellular N-acetylglutamic acid produced by these bacteria have three morphological effects on the white clover seedling roots: branching of root hairs, swelling of root tips, and increase in the number of cell divisions in undifferentiated cells found on the outer-most cell layer of the root. This suggests that N-acetylglutamic acid is involved in the stimulation of mitosis. The same effects were observed on the strawberry clover, but not in legumes. The effects of N-acetylglutamic acid on the clover species were more potent than the effects from glutamine, glutamate, arginine, or ammonia.[4]
Structure
N-Acetylglutamic acid is composed of two carboxylic acid groups and an amide group protruding from the second carbon. The structure of N-acetylglutamic acid at physiological pH (7.4) has all carboxyl groups deprotonated.
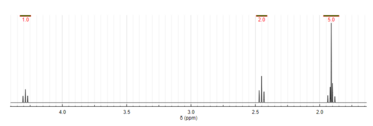
Proton NMR spectroscopy
The molecular structure of N-acetylglutamic acid was determined using proton NMR spectroscopy.[3] Proton NMR reveals the presence and functional group location of protons based on chemical shifts recorded on the spectrum.[11]
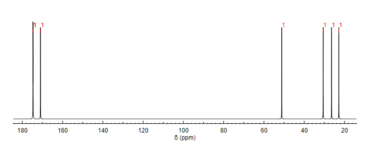
13C NMR spectroscopy
Like proton NMR, carbon-13 (13C) NMR spectroscopy is a method used in molecular structure determination. 13C NMR reveals the types of carbons present in a molecule based on chemical shifts that correspond to certain functional groups. N-Acetylglutamic acid exhibits carbonyl carbons most distinctly due to the three carbonyl-containing substituents.[12]
See also
- Glutamate
- Glutamic acid
- Urea cycle
- Arginine
- Ornithine
References
- ↑ "N-Acetyl-DL-glutamic acid - Compound Summary". PubChem Compound. USA: National Center for Biotechnology Information. 25 March 2005. Identification. https://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=185.
- ↑ Pubchem. "N-Acetyl L-glutamic acid" (in en). https://pubchem.ncbi.nlm.nih.gov/compound/N-Acetyl-L-glutamic_acid.
- ↑ 3.0 3.1 "N-Acetylglutamic acid: an extracellular nod signal of Rhizobium trifolii ANU843 that induces root hair branching and nodule-like primordia in white clover roots". The Journal of Biological Chemistry 266 (25): 16854–8. September 1991. doi:10.1016/S0021-9258(18)55380-1. PMID 1885611. http://www.jbc.org/content/266/25/16854.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 "N-Acetylglutamate and its changing role through evolution". The Biochemical Journal 372 (Pt 2): 279–90. June 2003. doi:10.1042/BJ20030002. PMID 12633501.
- ↑ 5.0 5.1 "Arginine biosynthesis in Escherichia coli: experimental perturbation and mathematical modeling". The Journal of Biological Chemistry 283 (10): 6347–58. March 2008. doi:10.1074/jbc.M705884200. PMID 18165237.
- ↑ Auditore, Joseph V.; Wade, Littleton; Olson, Erik J. (November 1966). "Occurrence of N-acetyl-L-glutamic Acid in the Human Brain" (in en). Journal of Neurochemistry 13 (11): 1149–1155. doi:10.1111/j.1471-4159.1966.tb04272.x. ISSN 0022-3042. PMID 5924663.
- ↑ "N-Acetyl-glutamic acid: evaluation of acute and 28-day repeated dose oral toxicity and genotoxicity". Food and Chemical Toxicology 47 (11): 2723–9. November 2009. doi:10.1016/j.fct.2009.07.036. PMID 19654033.
- ↑ Pelley, John W. (2007). "Chapter 14: Purine, Pyrimidine, and Single-Carbon Metabolism". Elsevier's Integrated Biochemistry. Elsevier. pp. 117–122. doi:10.1016/b978-0-323-03410-4.50020-1. ISBN 978-0-323-03410-4.
- ↑ "N-Acetylglutamate and N-acetylaspartate in soybeans (Glycine max L.), maize (Zea mays L.), [corrected] and other foodstuffs". Journal of Agricultural and Food Chemistry 56 (19): 9121–6. October 2008. doi:10.1021/jf801523c. PMID 18781757.
- ↑ "The course of phosphorus in the reaction of N-acetyl-L-glutamate kinase, determined from the structures of crystalline complexes, including a complex with an AlF−4 transition state mimic". Journal of Molecular Biology 331 (1): 231–44. August 2003. doi:10.1016/S0022-2836(03)00716-2. PMID 12875848.
- ↑ "Predict 1H proton NMR spectra". http://www.nmrdb.org/new_predictor/index.shtml?v=v2.87.7.
- ↑ "Predict 13C carbon NMR spectra". http://www.nmrdb.org/13c/index.shtml?v=v2.87.7.
External links
 |