Biology:T7 RNA polymerase
| T7 RNA polymerase | |||||||
|---|---|---|---|---|---|---|---|
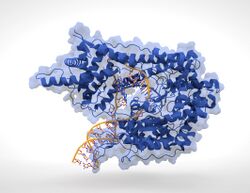 T7 RNA Polymerase (blue) producing mRNA (light-blue) from a double-stranded DNA template (orange). | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | 1 | ||||||
| PDB | 1MSW (ECOD) | ||||||
| UniProt | P00573 | ||||||
| |||||||
T7 RNA Polymerase is an RNA polymerase from the T7 bacteriophage that catalyzes the formation of RNA from DNA in the 5'→ 3' direction.[1]
Activity
T7 polymerase is extremely promoter-specific and transcribes only DNA downstream of a T7 promoter.[2] The T7 polymerase also requires a double stranded DNA template and Mg2+ ion as cofactor for the synthesis of RNA. It has a very low error rate. T7 polymerase has a molecular weight of 99 kDa.
Promoter
The promoter is recognized for binding and initiation of the transcription. The consensus in T7 and related phages is:[2]
5' * 3'
T7 TAATACGACTCACTATAGGGAGA
T3 AATTAACCCTCACTAAAGGGAGA
K11 AATTAGGGCACACTATAGGGAGA
SP6 ATTTACGACACACTATAGAAGAA
bind------------
-----------init
Transcription begins at the asterisk-marked guanine.[2]
Structure
T7 polymerase has been crystallised in several forms and the structures placed in the PDB. These explain how T7 polymerase binds to DNA and transcribes it. The N-terminal domain moves around as the elongation complex forms. The ssRNAP holds a DNA-RNA hybrid of 8bp.[3] A beta-hairpin specificity loop (residues 739-770 in T7) recognizes the promoter; swapping it out for one found in T3 RNAP makes the polymerase recognize T3 promoters instead.[2]
Similar to other viral nucleic acid polymerases, including T7 DNA polymerase from the same phage, the conserved C-terminal of T7 ssRNAP employs a fold whose organization has been likened to the shape of a right hand with three subdomains termed fingers, palm, and thumb.[4] The N-terminal is less conserved. It forms a promoter-binding domain (PBD) with helix bundles in phage ssRNAPs,[5] a feature not found in mitochondrial ssRNAPs.[6]
Related proteins
| DNA-directed RNA polymerase, phage-type | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | RNA_pol | ||||||||
| Pfam | PF00940 | ||||||||
| InterPro | IPR002092 | ||||||||
| SCOP2 | 1msw / SCOPe / SUPFAM | ||||||||
| |||||||||
T7 polymerase is a representative member of the single-subunit DNA-dependent RNAP (ssRNAP) family. Other members include phage T3 and SP6 RNA polymerases, the mitochondrial RNA polymerase (POLRMT), and the chloroplastic ssRNAP.[7][8] The ssRNAP family is structurally and evolutionarily distinct from the multi-subunit family of RNA polymerases (including bacterial and eukaryotic sub-families). In contrast to bacterial RNA polymerases, T7 polymerase is not inhibited by the antibiotic rifampicin. This family is related to single-subunit reverse transcriptase and DNA polymerase.[9]
Application
In biotechnology applications, T7 RNA polymerase is commonly used to transcribe DNA that has been cloned into vectors that have two (different) phage promoters (e.g., T7 and T3, or T7 and SP6) in opposite orientation. RNA can be selectively synthesized from either strand of the insert DNA with the different polymerases. The enzyme is stimulated by spermidine and in vitro activity is increased by the presence of carrier proteins (such as BSA).[10][11]
Homogeneously labeled single-stranded RNA can be generated with this system. Transcripts can be non-radioactively labeled to high specific activity with certain labeled nucleotides.
T7 RNA polymerase is used in the synthesis of mRNA and sgRNA.[12]
See also
References
- ↑ "T7 RNA Polymerase (20 U/µL)" (in en). https://www.thermofisher.com/order/catalog/product/EP0111.
- ↑ 2.0 2.1 2.2 2.3 "Promoter specificity determinants of T7 RNA polymerase". Proceedings of the National Academy of Sciences of the United States of America 95 (2): 515–9. January 1998. doi:10.1073/pnas.95.2.515. PMID 9435223. Bibcode: 1998PNAS...95..515R.
- ↑ "Structure of a T7 RNA polymerase elongation complex at 2.9 A resolution". Nature 420 (6911): 43–50. November 2002. doi:10.1038/nature01129. PMID 12422209.
- ↑ "Structure of the RNA-dependent RNA polymerase of poliovirus". Structure 5 (8): 1109–22. August 1997. doi:10.1016/S0969-2126(97)00261-X. PMID 9309225.
- ↑ "The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation". Science 322 (5901): 553–7. October 2008. doi:10.1126/science.1163433. PMID 18948533. Bibcode: 2008Sci...322..553D.
- ↑ "Structural Basis of Mitochondrial Transcription Initiation". Cell 171 (5): 1072–1081.e10. November 2017. doi:10.1016/j.cell.2017.10.036. PMID 29149603.
- ↑ "The phage RNA polymerases are related to DNA polymerases and reverse transcriptases". Molecular Microbiology 10 (1): 1–6. October 1993. doi:10.1111/j.1365-2958.1993.tb00897.x. PMID 7526118.
- ↑ "Mitochondrial and chloroplast phage-type RNA polymerases in Arabidopsis". Science 277 (5327): 809–11. August 1997. doi:10.1126/science.277.5327.809. PMID 9242608.
- ↑ "On the evolution of the single-subunit RNA polymerases". Journal of Molecular Evolution 45 (6): 671–81. December 1997. doi:10.1007/PL00006271. PMID 9419244. Bibcode: 1997JMolE..45..671C. https://www.researchgate.net/publication/13811504.
- ↑ "Characterization of T7-specific ribonucleic acid polymerase. 1. General properties of the enzymatic reaction and the template specificity of the enzyme". The Journal of Biological Chemistry 248 (6): 2235–44. March 1973. doi:10.1016/S0021-9258(19)44211-7. PMID 4570474.
- ↑ "Effects of solution conditions on the steady-state kinetics of initiation of transcription by T7 RNA polymerase". Biochemistry 33 (22): 6918–24. June 1994. doi:10.1021/bi00188a022. PMID 7911327.
- ↑ "T7 RNA Polymerase | NEB". https://www.neb.com/products/m0251-t7-rna-polymerase#Product%20Information.
Further reading
- "Structure and function in promoter escape by T7 RNA polymerase". Progress in Nucleic Acid Research and Molecular Biology 80: 323–47. 2005. doi:10.1016/S0079-6603(05)80008-X. ISBN 9780125400800. PMID 16164978.
- "T7 RNA polymerase". Progress in Nucleic Acid Research and Molecular Biology 73: 1–41. 2003. doi:10.1016/S0079-6603(03)01001-8. ISBN 9780125400732. PMID 12882513.
- "Structure and function of the bacteriophage T7 RNA polymerase (or, the virtues of simplicity)". Cellular & Molecular Biology Research 39 (4): 385–91. 1993. PMID 8312975.
- "Nuclease activity of T7 RNA polymerase and the heterogeneity of transcription elongation complexes". The Journal of Biological Chemistry 272 (13): 8644–52. March 1997. doi:10.1074/jbc.272.13.8644. PMID 9079696. - note that the nuclease activity reported here is an artifact.
External links
 |

