Biology:Shikimate kinase
| Shikimate kinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
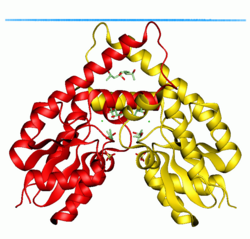 Shikimate kinase of Erwinia chrysanthemi | |||||||||
| Identifiers | |||||||||
| Symbol | SKI | ||||||||
| Pfam | PF01202 | ||||||||
| Pfam clan | CL0023 | ||||||||
| InterPro | IPR000623 | ||||||||
| PROSITE | PDOC00868 | ||||||||
| SCOP2 | 2shk / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 124 | ||||||||
| OPM protein | 1e6c | ||||||||
| |||||||||
| shikimate kinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
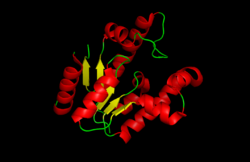 A cartoon representation of shikimate kinase from Mycobacterium tuberculosis. α-Helices are shown in red, the central β-sheet in yellow, and loops in green | |||||||||
| Identifiers | |||||||||
| EC number | 2.7.1.71 | ||||||||
| CAS number | 9031-51-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Shikimate kinase (EC 2.7.1.71) is an enzyme that catalyzes the ATP-dependent phosphorylation of shikimate to form shikimate 3-phosphate.[1] This reaction is the fifth step of the shikimate pathway,[2] which is used by plants and bacteria to synthesize the common precursor of aromatic amino acids and secondary metabolites. The systematic name of this enzyme class is ATP:shikimate 3-phosphotransferase. Other names in common use include shikimate kinase (phosphorylating), and shikimate kinase II.
Background
The shikimate pathway consists of seven enzymatic reactions by which phosphoenolpyruvate and erythrose 4-phosphate are converted to chorismate, the common precursor of the aromatic amino acids phenylalanine, tyrosine, and tryptophan. The aromatic amino acids are used in the synthesis of proteins and, in plants, fungi, and bacteria, give rise to a number of other specialized metabolites, such as phenylpropanoids and alkaloids. Chorismate and several other intermediates of the pathway serve as precursors for a number of other metabolites, such as folates, quinates, and quinones. The four enzymes that precede shikimate kinase in the pathway are DAHP synthase, 3-dehydroquinate synthase, 3-dehydroquinate dehydratase, and shikimate dehydrogenase, and the two that follow it are EPSP synthase and chorismate synthase. In fungi and protists, it is part of the AROM complex, in which the five central steps of the shikimate pathway are co-localized.[3] The pathway is not found in humans and other animals, which must obtain the aromatic amino acids from their food.
Activity
The reaction catalyzed by shikimate kinase is shown below:
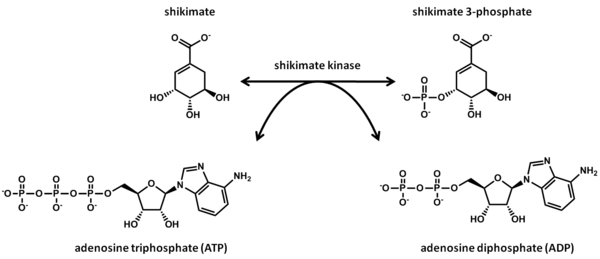

This reaction involves the transfer of a phosphate group from ATP to the 3-hydroxyl group of shikimate. Shikimate kinase thus has two substrates, shikimate and ATP, and two products, shikimate 3-phosphate and ADP.[4]
Examples
Human proteins containing this domain include: MAPK7 and THNSL1
References
- ↑ "Shikimate kinase isoenzymes in Salmonella typhimurium". The Journal of Biological Chemistry 243 (3): 676–7. February 1968. PMID 4866525.
- ↑ "The Shikimate Pathway". Annual Review of Plant Physiology and Plant Molecular Biology 50: 473–503. June 1999. doi:10.1146/annurev.arplant.50.1.473. PMID 15012217.
- ↑ "Architecture and functional dynamics of the pentafunctional AROM complex". Nature Chemical Biology 16 (9): 973–978. July 2020. doi:10.1038/s41589-020-0587-9. PMID 32632294.
- ↑ "Mechanism of phosphoryl transfer catalyzed by shikimate kinase from Mycobacterium tuberculosis". Journal of Molecular Biology 364 (3): 411–23. December 2006. doi:10.1016/j.jmb.2006.09.001. PMID 17020768.
 |
