Biology:Nucleobase
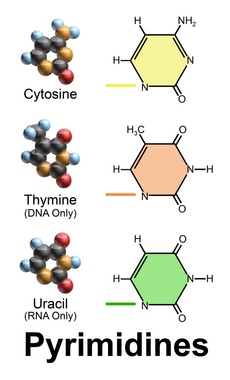
Nucleobases (nitrogenous bases or simply bases) are nitrogen-containing biological compounds that form nucleosides, which, in turn, are components of nucleotides, with all of these monomers constituting the basic building blocks of nucleic acids. The ability of nucleobases to form base pairs and to stack one upon another leads directly to long-chain helical structures such as ribonucleic acid (RNA) and deoxyribonucleic acid (DNA). Five nucleobases—adenine (A), cytosine (C), guanine (G), thymine (T), and uracil (U)—are called primary or canonical. They function as the fundamental units of the genetic code, with the bases A, G, C, and T being found in DNA while A, G, C, and U are found in RNA. Thymine and uracil are distinguished by merely the presence or absence of a methyl group on the fifth carbon (C5) of these heterocyclic six-membered rings.[1][page needed] In addition, some viruses have aminoadenine (Z) instead of adenine. It differs in having an extra amine group, creating a more stable bond to thymine.[2]
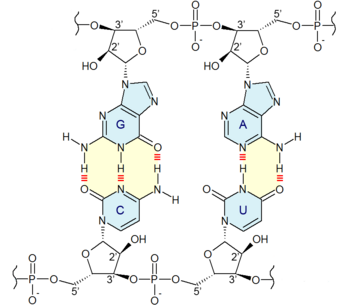
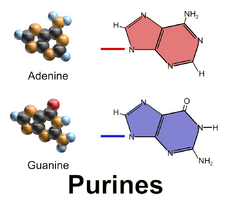
Adenine and guanine have a fused-ring skeletal structure derived of purine, hence they are called purine bases. The purine nitrogenous bases are characterized by their single amino group (–NH
2), at the C6 carbon in adenine and C2 in guanine.[3] Similarly, the simple-ring structure of cytosine, uracil, and thymine is derived of pyrimidine, so those three bases are called the pyrimidine bases.
Each of the base pairs in a typical double-helix DNA comprises a purine and a pyrimidine: either an A paired with a T or a C paired with a G. These purine-pyrimidine pairs, which are called base complements, connect the two strands of the helix and are often compared to the rungs of a ladder. Only pairing purine with pyrimidine ensures a constant width for the DNA. The A–T pairing is based on two hydrogen bonds, while the C–G pairing is based on three. In both cases, the hydrogen bonds are between the amine and carbonyl groups on the complementary bases.
Nucleobases such as adenine, guanine, xanthine, hypoxanthine, purine, 2,6-diaminopurine, and 6,8-diaminopurine may have formed in outer space as well as on earth.[4][5][6]
The origin of the term base reflects these compounds' chemical properties in acid–base reactions, but those properties are not especially important for understanding most of the biological functions of nucleobases.
Structure
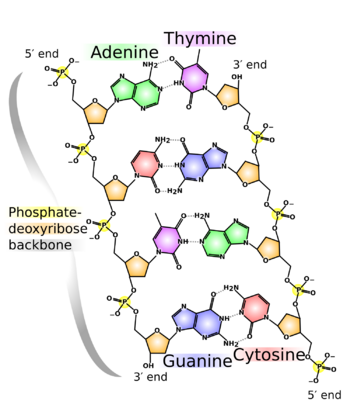
At the sides of nucleic acid structure, phosphate molecules successively connect the two sugar-rings of two adjacent nucleotide monomers, thereby creating a long chain biomolecule. These chain-joins of phosphates with sugars (ribose or deoxyribose) create the "backbone" strands for a single- or double helix biomolecule. In the double helix of DNA, the two strands are oriented chemically in opposite directions, which permits base pairing by providing complementarity between the two bases, and which is essential for replication of or transcription of the encoded information found in DNA.
Modified nucleobases
DNA and RNA also contain other (non-primary) bases that have been modified after the nucleic acid chain has been formed. In DNA, the most common modified base is 5-methylcytosine (m5C). In RNA, there are many modified bases, including those contained in the nucleosides pseudouridine (Ψ), dihydrouridine (D), inosine (I), and 7-methylguanosine (m7G).[7][8]
Hypoxanthine and xanthine are two of the many bases created through mutagen presence, both of them through deamination (replacement of the amine-group with a carbonyl-group). Hypoxanthine is produced from adenine, xanthine from guanine,[9] and uracil results from deamination of cytosine.
Modified purine nucleobases
These are examples of modified adenosine or guanosine.
| Nucleobase | 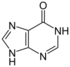 Hypoxanthine |
Chemical structure of xanthine Xanthine |
Chemical structure of 7-methylguanine 7-Methylguanine |
| Nucleoside | 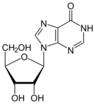 Inosine I |
Chemical structure of xanthosine Xanthosine X |
Chemical structure of 7-methylguanosine 7-Methylguanosine m7G |
Modified pyrimidine nucleobases
These are examples of modified cytidine, thymidine or uridine.
| Nucleobase | 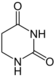 5,6-Dihydrouracil |
Chemical structure of 5-methylcytosine 5-Methylcytosine |
Chemical structure of 5-hydroxymethylcytosine 5-Hydroxymethylcytosine |
| Nucleoside | 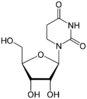 Dihydrouridine D |
Chemical structure of 5-methylcytidine 5-Methylcytidine m5C |
87px Pseudouridine |
Artificial nucleobases
A vast number of nucleobase analogues exist. The most common applications are used as fluorescent probes, either directly or indirectly, such as aminoallyl nucleotide, which are used to label cRNA or cDNA in microarrays. Several groups are working on alternative "extra" base pairs to extend the genetic code, such as isoguanine and isocytosine or the fluorescent 2-amino-6-(2-thienyl)purine and pyrrole-2-carbaldehyde.[10][11]
In medicine, several nucleoside analogues are used as anticancer and antiviral agents. The viral polymerase incorporates these compounds with non-canonical bases. These compounds are activated in the cells by being converted into nucleotides; they are administered as nucleosides as charged nucleotides cannot easily cross cell membranes.[citation needed] At least one set of new base pairs has been announced as of May 2014.[12]
Prebiotic condensation of nucleobases with ribose
In order to understand how life arose knowledge is required of chemical pathways that permit formation of the key building blocks of life under plausible prebiotic conditions. According to the RNA world hypothesis free-floating ribonucleotides were present in the primordial soup. These were the fundamental molecules that combined in series to form RNA. Molecules as complex as RNA must have arisen from small molecules whose reactivity was governed by physico-chemical processes. RNA is composed of purine and pyrimidine nucleotides, both of which are necessary for reliable information transfer, and thus Darwinian evolution. Nam et al.[13] demonstrated the direct condensation of nucleobases with ribose to give ribonucleosides in aqueous microdroplets, a key step leading to RNA formation. Similar results were obtained by Becker et al.[14]
See also
- Biology:Nucleoside – Any of several glycosylamines comprising a nucleobase and a sugar molecule
- Biology:Nucleotide – Biological molecules constituting nucleic acids
- Biology:Nucleic acid notation – Universal notation using the Roman characters A, C, G, and T to call the four DNA nucleotides
- Biology:Nucleic acid sequence – Succession of nucleotides in a nucleic acid
References
- ↑ Soukup, Garrett A. (2003). "Nucleic Acids: General Properties" (in en). eLS. American Cancer Society. doi:10.1038/npg.els.0001335. ISBN 9780470015902.
- ↑ "Some viruses thwart bacterial defenses with a unique genetic alphabet". 5 May 2021. https://www.sciencenews.org/article/virus-dna-z-bacteriophage-genetic-alphabet-bond-life.
- ↑ "Section 25.2, Purine Bases Can Be Synthesized de Novo or Recycled by Salvage Pathways.". Biochemistry. 5th Edition. https://www.ncbi.nlm.nih.gov/books/NBK22385/. Retrieved 2019-12-11.
- ↑ "Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases". Proceedings of the National Academy of Sciences of the United States of America (PNAS) 108 (34): 13995–8. August 2011. doi:10.1073/pnas.1106493108. PMID 21836052. Bibcode: 2011PNAS..10813995C.
- ↑ Steigerwald, John (8 August 2011). "NASA Researchers: DNA Building Blocks Can Be Made in Space". NASA. http://www.nasa.gov/topics/solarsystem/features/dna-meteorites.html.
- ↑ ScienceDaily Staff (9 August 2011). "DNA Building Blocks Can Be Made in Space, NASA Evidence Suggests". https://www.sciencedaily.com/releases/2011/08/110808220659.htm.
- ↑ Stavely, Brian E.. "BIOL2060: Translation". https://www.mun.ca/biology/desmid/brian/BIOL2060/BIOL2060-22/CB22.html.
- ↑ "Role of 5' mRNA and 5' U snRNA cap structures in regulation of gene expression" – Research – Retrieved 13 December 2010.
- ↑ "DNA damage and mutation in human cells exposed to nitric oxide in vitro". Proceedings of the National Academy of Sciences of the United States of America 89 (7): 3030–4. April 1992. doi:10.1073/pnas.89.7.3030. PMID 1557408. Bibcode: 1992PNAS...89.3030N.
- ↑ "A third base pair for the polymerase chain reaction: inserting isoC and isoG". Nucleic Acids Research 32 (6): 1937–41. 2004. doi:10.1093/nar/gkh522. PMID 15051811.
- ↑ "Fluorescent probing for RNA molecules by an unnatural base-pair system". Nucleic Acids Research 35 (16): 5360–69. 2007. doi:10.1093/nar/gkm508. PMID 17693436.
- ↑ "A semi-synthetic organism with an expanded genetic alphabet". Nature 509 (7500): 385–8. May 2014. doi:10.1038/nature13314. PMID 24805238. Bibcode: 2014Natur.509..385M.
- ↑ Nam, Inho; Nam, Hong Gil; Zare, Richard N. (2018). "Abiotic synthesis of purine and pyrimidine ribonucleosides in aqueous microdroplets". Proceedings of the National Academy of Sciences 115 (1): 36–40. doi:10.1073/pnas.1718559115. PMID 29255025. Bibcode: 2018PNAS..115...36N.
- ↑ Becker, Sidney; Feldmann, Jonas; Wiedemann, Stefan; Okamura, Hidenori; Schneider, Christina; Iwan, Katharina; Crisp, Antony; Rossa, Martin et al. (2019). "Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides". Science 366 (6461): 76–82. doi:10.1126/science.aax2747. PMID 31604305. Bibcode: 2019Sci...366...76B. https://epub.ub.uni-muenchen.de/71503/1/Science_Becker_2019.pdf.
External links
 |