Chemistry:Wybutosine
| |
| Names | |
|---|---|
| IUPAC name
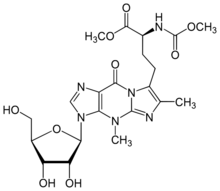
Methyl (2S)-4-(3,4′′-dimethyl-3H-imidazo[1′′,2′′:1,2]inosin-5′′-yl)-2-[(methoxycarbonyl)amino]butanoate
| |
| Systematic IUPAC name
Methyl (2S)-4-{3-[(2R,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]-4,6-dimethyl-9-oxo-4,9-dihydro-3H-imidazo[1,2-a]purin-7-yl}-2-[(methoxycarbonyl)amino]butanoate | |
| Identifiers | |
3D model (JSmol)
|
|
| Abbreviations | yW |
| ChEBI | |
| ChemSpider | |
PubChem CID
|
|
| UNII | |
| |
| |
| Properties | |
| C21H28N6O9 | |
| Molar mass | 508.488 g·mol−1 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
In biochemistry, wybutosine (yW) is a heavily modified nucleoside of phenylalanine transfer RNA that stabilizes interactions between the codons and anti-codons during protein synthesis.[1][2] Ensuring accurate synthesis of protein is essential in maintaining health as defects in tRNA modifications are able to cause disease. In eukaryotic organisms, it is found only in position 37, 3'-adjacent to the anticodon, of phenylalanine tRNA. Wybutosine enables correct translation through the stabilization of the codon-anticodon base pairing during the decoding process.[3]
Biosynthetic pathway
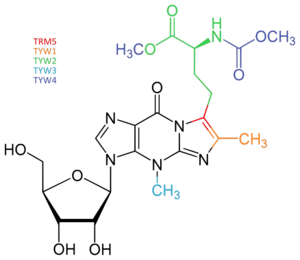
Using an S. cerevisiae model, the biosynthetic pathway of wybutosine was proposed. Proceeding through a multi-enzymatic process, the first step of the synthesis involves the enzyme N1-methyltransferase TRM5 which methylates the G37 site of phenylalanine tRNA and converts it to m1G37. Then m1G37 acts as a substrate for the enzyme TYW1 and, using pyruvate as a C-3 source, forms the tricyclic core of wybutosine with flavin mononucleotide (FMN) as a cofactor. The enzyme TYW2 then transfers the α-amino-α-carboxypropyl group from Ado-Met, a common substrate involved in methyl group transfers, to the lateral side chain at the C-7 position of yW-187 to form yW-86. TYW3 acts as a catalyst for N-4 methylation of yW-86 to produce yW-72. TYW4 and Ado-Met-dependent carboxymethyltransferase then methylates the α-carboxy group of yW-72 to give yW-57. Finally by a predicted second function of TYW4 or other unidentified factors, the methoxycarbonylation of the α-amino group of the yW-58 side chain gives wybutosine.[4]
Wybutosine has been chemically synthesized.[5][6]
Hypermodification and roles in RNA stabilization
Wybutosine and other unnatural nucleosides have been proposed to lead to a single outcome of hypermodification. This hypermodification at position 37 of tRNAPhe may allow for base- stacking interactions which play a key role in maintenance of the reading frame.[7] Through its large aromatic groups, stacking interactions with adjacent bases A36 and A38 are enhanced, which help to restrict the flexibility of the anticodon.[8] It has been found that when tRNAPhe lacks wybutosine, increased frameshifting occurs. Generally, modifications at position 37 prevent base pairing with neighboring nucleotides by helping to maintain and open the loop conformation, as well as generating an anticodon loop for decoding. The wybutosine modification of tRNAPhe is found to be conserved in archaea and eukarya but is not found in bacteria. Studies from the 1960s and 1970s noted that many mutations could lead to problems in translational accuracy. Further study of the mechanisms involved in translational accuracy revealed the importance of modifications on positions 34 and 37 of tRNA. Regardless of species, these sites of tRNA are almost always modified. The fact that wybutosine and its various derivatives are only found at position 37 may be indicative of the nature of the phenylalanine codons, UUU and UUC, and their predilection for ribosome slippage.[9] This has led to the assumption that tRNAPhe modification at position 37 correlates with the amount of polyuridine slippery sequences found in genomes.[10]
Frameshifting potential
Wybutosine's role in prevention of frameshifts has raised some questions into its importance, as there are other strategies beside modification with yW to prevent a shift. In Drosophila there is no modification at position 37 while in mammals yW is modified there. To explain this variability the idea of frameshifting potential has come about. This implies that cells use frameshifting as a mechanism to regulate themselves rather than trying to avoid frameshifting at all times.[11] It has been suggested that frameshifting may be used in a programmed manner, possibly to increase coding diversity.
References
- ↑ "Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA". EMBO J. 25 (10): 2142–54. 2006. doi:10.1038/sj.emboj.7601105. PMID 16642040.
- ↑ Perche-Letuvée, Phanélie; Molle, Thibaut; Forouhar, Farhad; Mulliez, Etienne; Atta, Mohamed (2 December 2014). "Wybutosine biosynthesis: Structural and mechanistic overview". RNA Biology 11 (12): 1508–1518. doi:10.4161/15476286.2014.992271. PMID 25629788.
- ↑ Suzuki, Yoko; Noma, Akiko; Suzuki, Tsutomu; Senda, Miki; Senda, Toshiya; Ishitani, Ryuichiro; Nureki, Osamu (October 2007). "Crystal Structure of the Radical SAM Enzyme Catalyzing Tricyclic Modified Base Formation in tRNA". Journal of Molecular Biology 372 (5): 1204–1214. doi:10.1016/j.jmb.2007.07.024. PMID 17727881.
- ↑ Jump up to: 4.0 4.1 Young, Anthony P.; Bandarian, Vahe (2018). "TYW1: A Radical SAM Enzyme Involved in the Biosynthesis of Wybutosine Bases". Radical SAM Enzymes. Methods in Enzymology. 606. pp. 119–153. doi:10.1016/bs.mie.2018.04.024. ISBN 978-0-12-812794-0.
- ↑ "Practical synthesis of wybutosine, the hypermodified nucleoside of yeast phenylalanine transfer ribonucleic acid". Chem. Pharm. Bull. 50 (4): 530–3. 2002. doi:10.1248/cpb.50.530. PMID 11964003.
- ↑ "Total synthesis of the hypermodified RNA bases wybutosine and hydroxywybutosine and their quantification together with other modified RNA bases in plant materials". Chemistry: A European Journal 19 (13): 4244–8. 2013. doi:10.1002/chem.201204209. PMID 23417961.
- ↑ Helm, M; Alfonzo, JD (2014). "Posttranscriptional RNA Modifications: playing metabolic games in a cell's chemical Legoland". Chem. Biol. 21 (2): 174–85. doi:10.1016/j.chembiol.2013.10.015. PMID 24315934.
- ↑ Stuart, JW; Koshlap, KM; Guenther, R; Agris, PF (2003). "Naturally-occurring modification restricts the anticodon domain conformational space of tRNA(Phe)". J Mol Biol 334 (5): 901–18. doi:10.1016/j.jmb.2003.09.058. PMID 14643656.
- ↑ Christian, T; Lahoud, G; Liu, C; Hou, YM (2010). "Control of catalytic cycle by a pair of analogous tRNA modification enzymes". J Mol Biol 400 (2): 204–17. doi:10.1016/j.jmb.2010.05.003. PMID 20452364.
- ↑ Jackman, JE; Alfonzo, JD (2013). "Transfer RNA modifications: nature's combinatorial chemistry playground". Wiley Interdiscip Rev RNA 4 (1): 35–48. doi:10.1002/wrna.1144. PMID 23139145.
- ↑ Waas, William F.; Druzina, Zhanna; Hanan, Melanie; Schimmel, Paul (September 2007). "Role of a tRNA Base Modification and Its Precursors in Frameshifting in Eukaryotes". Journal of Biological Chemistry 282 (36): 26026–26034. doi:10.1074/jbc.m703391200. PMID 17623669.
 |

