Biology:Aralkylamine N-acetyltransferase
| Aralkylamine N-acetyltransferase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
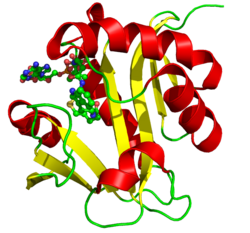 Crystallographic structure of aralkylamine N-acetyltransferase.[1] | |||||||||
| Identifiers | |||||||||
| EC number | 2.3.1.87 | ||||||||
| CAS number | 92941-56-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Aralkylamine N-acetyltransferase | |
|---|---|
| Identifiers | |
| Symbol | AANAT |
| NCBI gene | 15 |
| HGNC | 19 |
| OMIM | 600950 |
| RefSeq | NM_001088 |
| UniProt | Q16613 |
| Other data | |
| EC number | 2.3.1.87 |
| Locus | Chr. 17 q25 |
Aralkylamine N-acetyltransferase (AANAT) (EC 2.3.1.87), also known as arylalkylamine N-acetyltransferase or serotonin N-acetyltransferase (SNAT), is an enzyme that is involved in the day/night rhythmic production of melatonin, by modification of serotonin. It is in humans encoded by the ~2.5 kb AANAT gene[2] containing four exons, located on chromosome 17q25.[3] The gene is translated into a 23 kDa large enzyme. It is well conserved through evolution and the human form of the protein is 80 percent identical to sheep and rat AANAT. It is an acetyl-CoA-dependent enzyme of the GCN5-related family of N-acetyltransferases (GNATs). It may contribute to multifactorial genetic diseases such as altered behavior in sleep/wake cycle[2] and research is on-going with the aim of developing drugs that regulate AANAT function.
Nomenclature
The systematic name of this enzyme class is acetyl-CoA:2-arylethylamine N-acetyltransferase. Other names in common use include:
- AANAT
- Arylalkylamine N-acetyltransferase
- Melatonin rhythm enzyme
- Serotonin acetylase
- Serotonin acetyltransferase
- Serotonin N-acetyltransferase
The officially accepted name is aralkylamine N-acetyltransferase.[4]
Function and mechanism
Tissue distribution
The AANAT mRNA transcript is mainly expressed in the central nervous system (CNS). It is detectable at low levels in several brain regions including the pituitary gland as well as in the retina. It is most highly abundant in the pineal gland which is the site of melatonin synthesis. Brain and pituitary AANAT may be involved in the modulation of serotonin-dependent aspects of human behavior and pituitary function.[3]
Physiological function
In the pinealocyte cells of the pineal gland, aralkylamine N-acetyltransferase is involved in the conversion of serotonin to melatonin. It is the penultimate enzyme in the melatonin synthesis controlling the night/day rhythm in melatonin production in the vertebrate pineal gland. Melatonin is essential for seasonal reproduction, modulates the function of the circadian clock in the suprachiasmatic nucleus, and influences activity and sleep. Due to its important role in circadian rhythm, AANAT is subjected to extensive regulation that is responsive to light exposure (see Regulation). It may contribute to multifactorial genetic diseases such as altered behavior in sleep/wake cycle and mood disorders.[2]
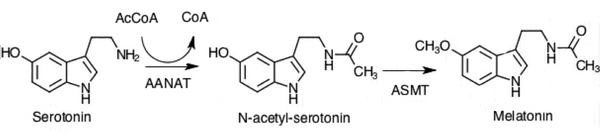
The chemical reactions catalyzed by AANAT
The primary chemical reaction that is catalyzed by aralkylamine N-acetyltransferase uses two substrates, acetyl-CoA and serotonin. AANAT catalyzes the transfer of the acetyl group of Acetyl-CoA to the primary amine of serotonin, thereby producing CoA and N-acetylserotonin. In humans, other endogenous substrates of the enzyme include specific trace amine neuromodulators, namely phenethylamine, tyramine, and tryptamine, in turn forming N-acetylphenethylamine, N-acetyltyramine, and N-acetyltryptamine.[5]
In the biosynthesis of melatonin, N-acetylserotonin is further methylated by another enzyme, N-acetylserotonin O-methyltransferase (ASMT) to generate melatonin. The N-acetyltransferase reaction has been suggested to be the rate-determining step, and thus Serotonin N-acetyltransferase has emerged as a target for inhibitor design (see below).[6]
AANAT obeys an ordered ternary-complex mechanism. The substrates bind sequentially (ordered) with acetyl-CoA binding to the free enzyme followed by the binding of serotonin to form the ternary complex. After the transfer of the acetyl group has occurred, the products are orderly released with N-acetyl-serotonin first and CoA last.[7]
Structure
Arylkylamine N-acetyltransferase is a monomeric polypeptide with a length of 207 amino acid residues, and with a molecular weight of 23,344 daltons. The secondary structure consists of alpha helices and beta sheets. It is 28 percent helical (10 helices; 60 residues) and 23 percent beta sheet (9 strands; 48 residues). This family shares four conserved sequence motifs designated A-D. Motif B serves as the location of the serotonin binding slot. The structure was determined by X-ray diffraction.[1]
Several structures have been solved for this class of enzymes, with PDB accession codes 1CJW,[8] 1B6B,[9] 1L0C,[1][10] and 1KUV/1KUX/1KUY.[1]
Aralkylamine N-acetyltransferase has also been crystallized in complex with 14-3-3ζ from the 14-3-3 protein family, with the PDB accession code 1IB1.[11]
The GNAT superfamily
Aralkylamine N-acetyltransferase belongs to the GCN5-related N-acetyltransferase (GNAT) superfamily which consists 10,000 acetyltransferases, named so because of their sequence homology to a class of eukaryotic transcription factors, therein the yeast GCN5. Other well-studied members of the superfamily are glucosamine-6-phosphate N-acetyltransferase and histone acetyltransferases.
All members of this superfamily has a structurally conserved fold consisting of an N-terminal strand followed by two helices, three antiparallel β-strands, followed by a ‘‘signature’’ central helix, a fifth β-strand, a fourth α-helix and a final β-strand. These elements are nearly universally conserved in spite of poor pairwise identity in sequence alignments.[12]
Regulation
Regulation of AANAT varies between species. In some, AANAT levels oscillate dramatically between light and dark periods, and thus control melatonin synthesis. In others, rhythm is regulated primarily on the protein level.[13] One example is in rodents, where AANAT mRNA levels increase more than 100-fold in dark periods. In other species, cyclic AMP plays an important part in inhibition of proteolytic degradation of AANAT, elevating protein levels at night. Experiments using human AANAT expressed in a 1E7 cell line show an ~8-fold increase in enzyme activity upon exposure to forskolin.[14]
Dynamic degradation of AANAT mRNA has proven essential to the circadian action of the enzyme. The 3’UTR sequences have importance with regards to the rhythmic degradation of AANAT mRNA in some species. In rodents, various hnRNPs maintain dynamic degradation of AANAT mRNA. In other species, such as ungulates and primates, the stable AANAT mRNAs with a shorter 3’UTR is suspected not to be under control of the hnRNPs that bind and direct degradation of AANAT mRNA in rodents.[15]
Exposure to light induces signals to travel from retinal cells, ultimately causing a drop in norepinephrine stimulation of the pineal gland. This, in turn, leads to a signaling cascade, resulting in Protein Kinase A phosphorylation of two key Ser and Thr residues of serotonin N-acetyltransferase. Phosphorylation of these residues causes changes in catalytic activity through recruitment and interaction with 14-3-3 proteins, specifically 14-3-3ζ.[16]
Another protein which interacts and regulates AANAT activity is protein kinase C. Protein kinase C acts, like protein kinase A, on threonine and serine residues, enhancing the stability and enzymatic activity of AANAT.[17]
Inhibition of the acetyl-CoA-binding to the catalytic site through the formation and cleavage of intramolecular disulfide bonds has been suggested to be a mechanism of regulation. Formation of a disulfide bond between two cystein residues within the protein closes the hydrophobic funnel of the catalytic site, and thus acts as an on/off switch for catalytic activity. It is not yet certain if this mechanism is present in in vivo cells through the regulation of intracellular redox conditions, but it is suggested that glutathione (GSH) could be an in vivo regulator of the formation and cleavage of these disulfide bonds.[18]
AANAT inhibitors and clinical relevance
Inhibitors of AANAT may eventually lead to development of a drug that would be useful in circadian biology research and in the treatment of sleep and mood disorders. Synthetic inhibitors of the enzyme have been discovered.[19][20][21] However, no AANAT inhibitor with potent in vivo activity has been reported.[22] Up to now, five classes of AANAT inhibitors have been described in the literature.[6] Below are the five classes:
Melatonin derivatives
Since it was reported that melatonin is a competitive inhibitor of AANAT, this neurotransmitter seems to exert an autoregulatory control on its own biosynthesis. Thus, loose structural analogues of the indolamine hormone were evaluated on AANAT, and moderate inhibitors were discovered.[23]
Peptidic inhibitors
Peptide combinatorial libraries of tri-, tetra-, and pentapeptides with various amino acid compositions were screened as potential sources of inhibitors, to see if it serves as either pure or mixed competitive inhibitor for the hAANAT enzyme. Molecular modeling and structure-activity relationship studies made it possible to pinpoint the amino acid residue of the pentapeptide inhibitor S 34461 that interacts with the cosubstrate-binding site.[24]
Bisubstrate analogs
It is suggested that AANAT catalyzes the transfer of an acetyl group from acetyl-CoA to serotonin, with the involvement of an intermediate ternary complex, to produce N-acetylserotonin. Based on this mechanism, it might be expected that a bisubstrate analog inhibitor, derived from the tethering of indole and CoASH parts, could potentially mimic the ternary complex and exert strong inhibition of AANAT.[25] The first bisubstrate analog (1), which links tryptamine and CoA via an acetyl bridge, was synthesized by Khalil and Cole, and shown to be a very potent and specific AANAT inhibitor.[26]
N-Haloacetylated derivatives
AANAT has shown that it also has a secondary alkyltransferase activity as well as acetyltransferase activity.[27] N-Haloacetyltryptamines were developed and serve as substrates of AANAT alkyltransferase and are also potent (low micromolar) in vitro inhibitors against AANAT acetyltransferase activity. AANAT catalyzes reaction between N-bromoacetyltryptamine (BAT) and reduced CoA, resulting a tight-binding bisubstrate analog inhibitor.[27][28] The first synthesized cell-permeable inhibitor of AANAT N-bromoacetyltryptamine was studied further on melatonin secretion from rat and pig pineal glands.[29] New N-halogenoacetyl derivatives leading to a strong in situ inhibition of AANAT. The concept behind the mechanism of action of these precursors was studied by following the biosynthesis of the inhibitor from tritiated-BAT in a living cell.[20]
Rhodanine-based compounds
The first druglike and selective inhibitors of AANAT has been identified. Lawrence M. Szewczuk et al. have virtually screened more than a million compounds by 3D high-throughput docking into the active site of X-ray structure for AANAT, and then tested 241 compounds as inhibitors. One compound class which containing a rhodanine scaffold has shown low micromolar competitive inhibition against acetyl-CoA and proved to be effective in blocking melatonin production in pineal cells.[19]
The recent study about inhibitor of AANAT has described the discovery of a new class of nonpeptidic AANAT inhibitors based on a 2,2′-bithienyl scaffold.[22]
See also
References
- ↑ 1.0 1.1 1.2 1.3 PDB: 1KUX; "X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition". J. Mol. Biol. 317 (2): 215–24. March 2002. doi:10.1006/jmbi.2001.5371. PMID 11902838.
- ↑ 2.0 2.1 2.2 "Entrez Gene: arylalkylamine N-acetyltransferase". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=15.
- ↑ 3.0 3.1 "The human serotonin N-acetyltransferase (EC 2.3.1.87) gene (AANAT): structure, chromosomal localization, and tissue expression". Genomics 34 (1): 76–84. May 1996. doi:10.1006/geno.1996.0243. PMID 8661026.
- ↑ "IUBMB Enzyme Nomenclature EC 2.3.1.87". Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB). http://www.chem.qmul.ac.uk/iubmb/enzyme/EC2/3/1/87.html.
- ↑ "EC 2.3.1.87 - aralkylamine N-acetyltransferase". Technische Universität Braunschweig. July 2014. http://www.brenda-enzymes.org/enzyme.php?ecno=2.3.1.87&Suchword=&organism%5B%5D=Homo+sapiens&show_tm=0.
- ↑ 6.0 6.1 "Serotonin N-acetyltransferase: mechanism and inhibition". Curr. Med. Chem. 9 (12): 1187–99. June 2002. doi:10.2174/0929867023370013. PMID 12052171.
- ↑ J. De Angelis; J. Gastel; D. C. Klein; P. A. Cole (January 1998). "Kinetic analysis of the catalytic mechanism of serotonin N-acetyltransferase (EC 2.3.1.87)". The Journal of Biological Chemistry 273 (5): 3045–3050. doi:10.1074/jbc.273.5.3045. PMID 9446620.
- ↑ "The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 A resolution with a bisubstrate analog". Cell 97 (3): 361–9. April 1999. doi:10.1016/S0092-8674(00)80745-X. PMID 10319816.
- ↑ "Melatonin biosynthesis: the structure of serotonin N-acetyltransferase at 2.5 A resolution suggests a catalytic mechanism". Mol. Cell 3 (1): 23–32. January 1999. doi:10.1016/S1097-2765(00)80171-9. PMID 10024876.
- ↑ "Investigation of the roles of catalytic residues in serotonin N-acetyltransferase". J. Biol. Chem. 277 (20): 18118–26. May 2002. doi:10.1074/jbc.M200595200. PMID 11884405.
- ↑ "Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. a role for scaffolding in enzyme regulation". Cell 105 (2): 257–67. April 2001. doi:10.1016/S0092-8674(01)00316-6. PMID 11336675.
- ↑ Matthew W. Vetting; Luiz Pedro S. de Carvalho; Michael Yu; Subray S. Hegde; Sophie Magnet; Steven L. Roderick; John S. Blanchard (January 2005). "Structure and functions of the GNAT superfamily of acetyltransferases". Archives of Biochemistry and Biophysics 433 (1): 212–226. doi:10.1016/j.abb.2004.09.003. PMID 15581578.
- ↑ Klein, D. C.; Coon, S. L.; Roseboom, P. H.; Weller, J. L.; Bernard, M.; Gastel, J. A.; Zatz, M.; Iuvone, P. M. et al. (1997). "The melatonin rhythm-generating enzyme: Molecular regulation of serotonin N-acetyltransferase in the pineal gland". Recent Progress in Hormone Research 52: 307–357; discussion 357–8. PMID 9238858.
- ↑ "cAmp regulation of arylalkylamine N-acetyltransferase (AANAT, EC 2.3.1.87): a new cell line (1E7) provides evidence of intracellular AANAT activation". J. Biol. Chem. 276 (26): 24097–107. June 2001. doi:10.1074/jbc.M011298200. PMID 11313340.
- ↑ "Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation". Mol. Cell. Biol. 25 (8): 3232–46. April 2005. doi:10.1128/MCB.25.8.3232-3246.2005. PMID 15798208.
- ↑ "Analysis of serotonin N-acetyltransferase regulation in vitro and in live cells using protein semisynthesis". Biochemistry 47 (39): 10407–19. September 2008. doi:10.1021/bi801189d. PMID 18771288.
- ↑ "Protein kinase C regulates the activity and stability of serotonin N-acetyltransferase". J. Neurochem. 90 (2): 442–54. July 2004. doi:10.1111/j.1471-4159.2004.02495.x. PMID 15228600.
- ↑ "An intramolecular disulfide bridge as a catalytic switch for serotonin N-acetyltransferase". J. Biol. Chem. 277 (46): 44229–35. November 2002. doi:10.1074/jbc.M203305200. PMID 12215431.
- ↑ 19.0 19.1 Lawrence M. Szewczuk; S. Adrian Saldanha; Surajit Ganguly; Erin M. Bowers; Margarita Javoroncov; Balasubramanyam Karanam; Jeffrey C. Culhane; Marc A. Holbert et al. (November 2007). "De novo discovery of serotonin N-acetyltransferase inhibitors". Journal of Medicinal Chemistry 50 (22): 5330–5338. doi:10.1021/jm0706463. PMID 17924613.
- ↑ 20.0 20.1 "New substrate analogues of human serotonin N-acetyltransferase produce in situ specific and potent inhibitors". Eur. J. Biochem. 271 (2): 418–28. January 2004. doi:10.1046/j.1432-1033.2003.03942.x. PMID 14717709.
- ↑ "Novel bisubstrate analog inhibitors of serotonin N-acetyltransferase: the importance of being neutral". Bioorg. Chem. 31 (5): 398–411. October 2003. doi:10.1016/S0045-2068(03)00081-6. PMID 12941292.
- ↑ 22.0 22.1 "Receptor- and ligand-based study on novel 2,2'-bithienyl derivatives as non-peptidic AANAT inhibitors". J Chem Inf Model 50 (3): 446–60. March 2010. doi:10.1021/ci9004805. PMID 20196559.
- ↑ "Structure-activity relationships for substrates and inhibitors of pineal 5-hydroxytryptamine-N-acetyltransferase: preliminary studies". Eur. J. Pharmacol. 307 (2): 133–40. June 1996. doi:10.1016/0014-2999(96)00228-2. PMID 8832214.
- ↑ "Substrate specificity and inhibition studies of human serotonin N-acetyltransferase". J. Biol. Chem. 275 (12): 8794–805. March 2000. doi:10.1074/jbc.275.12.8794. PMID 10722724.
- ↑ Page, A.I. (1990). "Enzyme inhibition". Comprehensive Medicinal Chemistry 2: 61–87.
- ↑ Khalil, Ehab M.; Philip A. Cole (6 June 1998). "A Potent Inhibitor of the Melatonin Rhythm Enzyme". J. Am. Chem. Soc. 120 (24): 6195–6196. doi:10.1021/ja981365a.
- ↑ 27.0 27.1 "Mechanism-based inhibition of the melatonin rhythm enzyme: pharmacologic exploitation of active site functional plasticity". Proceedings of the National Academy of Sciences of the United States of America 96 (22): 12418–12423. October 1999. doi:10.1073/pnas.96.22.12418. PMID 10535937. Bibcode: 1999PNAS...9612418K.
- ↑ "Mechanistic studies on the alkyltransferase activity of serotonin N-acetyltransferase". Chemistry & Biology 8 (4): 379–389. April 2001. doi:10.1016/s1074-5521(01)00020-5. PMID 11325593.
- ↑ "N-bromoacetyltryptamine strongly and reversibly inhibits in vitro melatonin secretion from mammalian pinealocytes". Neuroendocrinology Letters 26 (5): 581–592. October 2005. PMID 16264397.
Further reading
- "Arylamine N-acetyltransferase and arylalkylamine N-acetyltransferase in the mammalian pineal gland". J. Biol. Chem. 259 (17): 10913–8. 1984. doi:10.1016/S0021-9258(18)90600-9. PMID 6469990.
- "Substrate specificity and inhibition studies of human serotonin N-acetyltransferase". J. Biol. Chem. 275 (12): 8794–805. 2000. doi:10.1074/jbc.275.12.8794. PMID 10722724.
- "A potent inhibitor of the melatonin rhythm enzyme". J. Am. Chem. Soc. 120 (24): 6195–6196. 1998. doi:10.1021/ja981365a.
External links
- Serotonin+N-Acetyltransferase at the US National Library of Medicine Medical Subject Headings (MeSH)
- AANAT human gene location in the UCSC Genome Browser.
- AANAT human gene details in the UCSC Genome Browser.
This article incorporates text from the United States National Library of Medicine, which is in the public domain.
 |