Biology:Cytochrome P450 aromatic O-demethylase
| Aromatic O-demethylase, cytochrome P450 subunit | |||||||
|---|---|---|---|---|---|---|---|
 | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | gcoA | ||||||
| PDB | 5NCB (ECOD) | ||||||
| UniProt | P0DPQ7 | ||||||
| Other data | |||||||
| EC number | 1.14.14.- | ||||||
| |||||||
| Aromatic O-demethylase, reductase subunit | |||||||
|---|---|---|---|---|---|---|---|
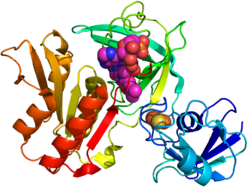 Crystal structure of gcoB (cartoon diagram) complexed with FAD (magenta spheres) and an iron–sulfur cluster (orange/yellow) based on PDB: 5OGX.[1] | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | gcoB | ||||||
| PDB | 5OGX (ECOD) | ||||||
| UniProt | P0DPQ8 | ||||||
| Other data | |||||||
| EC number | 1.6.2.- | ||||||
| |||||||
Cytochrome P450 aromatic O-demethylase is a bacterial enzyme that catalyzes the demethylation of lignin and various lignols. The net reaction follows the following stoichiometry, illustrated with a generic methoxy arene:[1]
- ArOCH3 + O2 + 2 e− + 2 H+ → ArOH + CH2O + H2O
The enzyme is notable for its promiscuity, affecting the O-demethylation of a range of substrates, including lignin.
It is a heterodimeric protein derived from the products of two genes. The component proteins are a cytochrome P450 enzyme (encoded by the gcoA gene from the family CYP255A) and a three-domain reductase (encoded by the gcoB gene) complexed with three cofactors (2Fe-2S, FAD, and NADH).[1]
Mechanism
GcoA and GcoB form a dimer complex in solution. GcoA process the substrate while GcoB provides the electrons to support the mixed function oxidase. As with other P450's, monooxygenation of the substrate proceeds concomitantly with reduction of half an equivalent of O2 to water. An oxygen rebound mechanism can be assumed. GcoA positions the aromatic ring within the hydrophobic active site cavity where the heme is located.[2][3]
Structure
GcoA has a typical P450 structure: a thiolate-ligated heme next to a buried active site. GcoB is however unusual. Cytochrome P450s normally are complemented by either a cytochrome P450 reductase[4] or a ferredoxin and ferredoxin reductase; its electrons are carried by NAD+ or NADP+. GcoB however has a single polypeptide. This polypeptide has an N-terminal ferredoxin with both an NAD(P)+ and also an FAD binding region.
CcoA and GcoB are closely interlinked, acting as an heterodimer in solution. The surface of GcoB has an acidic patch that must interact with the matching basic region in GcoA. It is assumed that the part of GcoB interacting with GcoA is at the intersection between the FAD binding domain and ferredoxin domain. To achieve this GcoB would have to go through some structural change, which would represent a new class of P450 systems (family N).[5][6][7]
Potential applications
Cytochrome P450 aromatic O-demethylase assists in the partial O-demethylation of lignin. The resulting 1,2-diols are well suited for oxidative degradation via intra- and extra-diol dioxygenases. Thus O-demethylated lignins are potentially susceptible to partial depolymerization.[8] With fewer crosslinks, the modified ligand is potentially more useful than the precursor.,[9] ranging from fuels[10][11]
References
- ↑ 1.0 1.1 1.2 1.3 "A promiscuous cytochrome P450 aromatic O-demethylase for lignin bioconversion". Nature Communications 9 (1): 2487. June 2018. doi:10.1038/s41467-018-04878-2. PMID 29950589. Bibcode: 2018NatCo...9.2487M.
- ↑ "The ins and outs of ring-cleaving dioxygenases". Critical Reviews in Biochemistry and Molecular Biology 41 (4): 241–67. 2006. doi:10.1080/10409230600817422. PMID 16849108.
- ↑ "Redox-linked domain movements in the catalytic cycle of cytochrome p450 reductase". Structure 21 (9): 1581–9. September 2013. doi:10.1016/j.str.2013.06.022. PMID 23911089.
- ↑ "Three-dimensional structure of NADPH-cytochrome P450 reductase: prototype for FMN- and FAD-containing enzymes". Proceedings of the National Academy of Sciences of the United States of America 94 (16): 8411–6. August 1997. doi:10.1073/pnas.94.16.8411. PMID 9237990. Bibcode: 1997PNAS...94.8411W.
- ↑ "Structure of a cytochrome P450-redox partner electron-transfer complex". Proceedings of the National Academy of Sciences of the United States of America 96 (5): 1863–8. March 1999. doi:10.1073/pnas.96.5.1863. PMID 10051560. Bibcode: 1999PNAS...96.1863S.
- ↑ "Structural basis for effector control and redox partner recognition in cytochrome P450". Science 340 (6137): 1227–30. June 2013. doi:10.1126/science.1235797. PMID 23744947. Bibcode: 2013Sci...340.1227T.
- ↑ "Structure and function of cytochromes P450: a comparative analysis of three crystal structures". Structure 3 (1): 41–62. January 1995. doi:10.1016/s0969-2126(01)00134-4. PMID 7743131.
- ↑ "Enzymatic conversion of lignin into renewable chemicals". Current Opinion in Chemical Biology 29: 10–7. December 2015. doi:10.1016/j.cbpa.2015.06.009. PMID 26121945.
- ↑ "Opportunities and challenges in biological lignin valorization". Current Opinion in Biotechnology 42: 40–53. December 2016. doi:10.1016/j.copbio.2016.02.030. PMID 26974563.
- ↑ "Adipic acid production from lignin". Energy & Environmental Science 8 (2): 617–628. 2015. doi:10.1039/c4ee03230f. ISSN 1754-5692.
- ↑ "Systems biology-guided biodesign of consolidated lignin conversion". Green Chemistry 18 (20): 5536–5547. 2016. doi:10.1039/c6gc01131d. ISSN 1463-9262.
 |




