Biology:Demethylase
Demethylases are enzymes that remove methyl (CH3) groups from nucleic acids, proteins (particularly histones), and other molecules. Demethylases are important epigenetic proteins, as they are responsible for transcriptional regulation of the genome by controlling the methylation of DNA and histones, and by extension, the chromatin state at specific gene loci.
Histone lysine demethylation
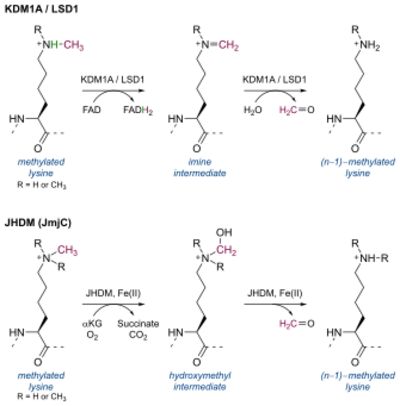
Histone methylation was initially considered an effectively irreversible process as the half-life of the histone methylation was approximately equal to the histone half-life.[1] Histone lysine demethylase LSD1 (later classified as KDM1A) was first identified in 2004 as a nuclear amine oxidase homolog.[2] Two main classes of histone lysine demethylases exist, defined by their mechanisms: flavin adenine dinucleotide (FAD)-dependent amine oxidases and α-ketoglutarate-dependent hydroxylases.
Histone lysine demethylases possess a variety of domains that are responsible for histone recognition, DNA binding, methylated amino acid substrate binding and catalytic activity. These include:
- FAD-dependent amine oxidase domains containing the active catalytic site of KDM1
- Jumonji-C domains containing the active catalytic site of KDM2 through KDM8[3][4]
- Jumonji-N domains responsible for Jumonji-C domain conformation stability
- SWIRM (SWI3P, RSC8P and Moira) domains proposed as an anchor site for histone substrates and responsible for chromatin stability
- PHD, CXXC and C5HC2 zinc finger domains responsible for histone recognition and binding
Histone lysine demethylases are classified according to their domains and unique substrate specificities. The lysine substrates and identified according to their position in the corresponding histone amino acid sequence and methylation state (for example, H3K9me3 refers to trimethylated histone 3 lysine 9.)


- KDM1
- The KDM1 homologs include KDM1A and KDM1B. KDM1A demethylates H3K4me1/2 and H3K9me1/2, and KDM1B emethylates H3K4me1/2. KDM1 activity is critical to embryogenesis and tissue-specific differentiation, as well as oocyte growth.[1] Deletion of the gene for KDM1A can have effects on the growth and differentiation of embryonic stem cells and is universally lethal in knockout mice.[5][6] KDM1A gene expression is observed to be upregulated in some cancers,[7][8] and so KDM1A inhibition has therefore been considered a possible epigenetic treatment for cancer.[9][10][11]:KDM1B, however, is mostly involved in oocyte development. Deletion of this gene leads to maternal effect lethality in mice.[12] Orthologs of KDM1 in D. melanogaster and C. elegans appear to function similarly to KDM1B rather than KDM1A.[13][14]
- KDM2
- The KDM2 homologs include KDM2A and KDM2B. KDM2A and KDM2B demethylate H3K4me3 and H3K36me2/3. KDM2A has roles in either promoting or inhibiting tumor function, and KDM2B has roles in oncogenesis.[1] KDM2A and KDM2B possess CXXC zinc finger domains responsible for binding to unmethylated CpG islands, and it is believed that they may bind to many gene regulatory elements in the absence of sequence-specific transcription factors.[15]:Overexpressed KDM2B has been observed in human lymphoma and adenocarcinoma, and underexpressed KDM2B has been observed in human prostate cancer and glioblastoma. KDM2B has been additionally shown to prevent senescence in some cells through ectopic expression.[16]
- KDM3
- The KDM3 homologs include KDM3A, KDM3B and KDM3C. KDM3A, KDM3B and KDM3C demethylate H3K9me1/2. KDM3A has roles in spermatogenesis and metabolic functions, however, the activity of KDM3B and KDM3C are not specifically known.[1]:Knockdown studies of KDM3A in mice resulted in male infertility and adult onset-obesity. Additional studies have indicated that KDM3A may play a role in regulation of androgen receptor-dependent genes as well as genes involved in pluripotency, indicating a potential role for KDM3A in tumorigenesis.[17]
- KDM4
- The KDM4 homologs include KDM4A, KDM4B, KDM4C, KDM4D, KDM4E and KDM4F. KDM4A, KDM4B and KDM4C demethylate H3K9me2/3, H3K9me3 and H3K36me2/3, and KDM4D, KDM4E and KDM4F demethylate H3K9me2/3. KDM4A, KDM4B, KDM4C and KDM4D have roles in tumorigenesis, however, the activity of KDM4E and KDM4F are not specifically known..[1] KDM4B upregulation has bee observed in medulloblastoma, and KDM4C amplification has been documented in oesophageal squamous carcinoma, medulloblastoma and breast cancer.[18][19][20][21] Other gene expression data has also suggested KDM4A, KDM4B, and KDM4C are overexpressed in prostate cancer.[22]
- KDM5
- The KDM5 homologs includes KDM5A, KDM5B, KDM5C and KDM5D. KDM5A, KDM5B, KDM5C and KDM5D demethylate H3K4me2/3.[1] The KDM5 family appears to regulate key developmental functions, including cellular differentiation, mitochondrial function and cell cycle progression.[23][24][25][26][27][28] KDM5B and KDM5C have also shown to interaction with PcG proteins, which are involved in transcriptional repression. KDM5C mutations on the X-chromosome have also been observed in patients with X-linked intellectual disability.[29] Depletion of KDM5C homologs in D. rerio have shown brain-patterning defects and neuronal cell death.[30]
- KDM6
- The KDM6 family includes KDM6A, KDM6B and KDM6C. KDM6A and KDM6B demethylate H3K27me2/3, and KDM4C demethylates H3K27me3. KDM6A and KDM6B possess tumor-suppressive characteristics. KDM6A knockdowns in fibroblasts lead to an immediate increase in fibroblast population. KDM6B expressed in fibroblasts induces oncogenes of the Ras/Raf/MEK/ERK pathway.[31] Point mutations of KDM6A have been identified as one cause of Kabuki syndrome, a congenital disorder resulting in intellectual disability.[32][33] Deletion of KDM6A in D. rerio results in decreased expression of HOX genes, which play a role in regulating body patterning during development.[34] In mammalian studies, KDM6A has been shown to regulate HOX genes as well.[35][36] Mutation of KDM5B disrupt gonad development in C.elegans.[35] Other studies have shown that KDM6B expression is upregulated in activated macrophages and dynamically expressed during differentiation of stem cells.[37][38]
Ester demethylation
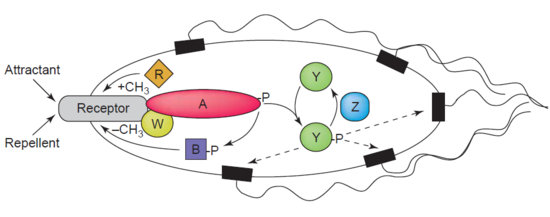
Another example of a demethylase is protein-glutamate methylesterase, also known as CheB protein (EC 3.1.1.61), which demethylates MCPs (methyl-accepting chemotaxis proteins) through hydrolysis of carboxylic ester bonds. The association of a chemotaxis receptor with an agonist leads to the phosphorylation of CheB. Phosphorylation of CheB protein enhances its catalytic MCP demethylating activity resulting in adaption of the cell to environmental stimuli.[39] MCPs respond to extracellular attractants and repellents in bacteria like E. coli in chemotaxis regulation. CheB is more specifically termed a methylesterase, as it removes methyl groups from methylglutamate residues located on the MCPs through hydrolysis, producing glutamate accompanied by the release of methanol.[40]
CheB is of particular interest to researchers as it may be a therapeutic target for mitigating the spread of bacterial infections.[41]
See also
- Chemotaxis
- Esterase
- Transferase
- Methylase
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 "Histone demethylases in development and disease". Trends in Cell Biology 20 (11): 662–71. Nov 2010. doi:10.1016/j.tcb.2010.08.011. PMID 20863703.
- ↑ "Histone demethylation mediated by the nuclear amine oxidase homolog LSD1". Cell 119 (7): 941–53. Dec 2004. doi:10.1016/j.cell.2004.12.012. PMID 15620353.
- ↑ "Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases". Annual Review of Biochemistry 79: 155–79. 2010. doi:10.1146/annurev.biochem.78.070907.103946. PMID 20373914.
- ↑ "The epigenetic modifier JMJD6 is amplified in mammary tumors and cooperates with c-Myc to enhance cellular transformation, tumor progression, and metastasis". Clin Epigenetics 8 (38): 38. Apr 2016. doi:10.1186/s13148-016-0205-6. PMID 27081402.
- ↑ "The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation". Nature Genetics 41 (1): 125–9. Jan 2009. doi:10.1038/ng.268. PMID 19098913.
- ↑ "Opposing LSD1 complexes function in developmental gene activation and repression programmes". Nature 446 (7138): 882–7. Apr 2007. doi:10.1038/nature05671. PMID 17392792. Bibcode: 2007Natur.446..882W.
- ↑ "Androgen receptor coactivators lysine-specific histone demethylase 1 and four and a half LIM domain protein 2 predict risk of prostate cancer recurrence". Cancer Research 66 (23): 11341–7. Dec 2006. doi:10.1158/0008-5472.CAN-06-1570. PMID 17145880.
- ↑ "Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology". Carcinogenesis 31 (3): 512–20. Mar 2010. doi:10.1093/carcin/bgp324. PMID 20042638.
- ↑ "LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription". Nature 437 (7057): 436–9. Sep 2005. doi:10.1038/nature04020. PMID 16079795. Bibcode: 2005Natur.437..436M.
- ↑ "Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy". Cancer Research 69 (5): 2065–71. Mar 2009. doi:10.1158/0008-5472.CAN-08-1735. PMID 19223552.
- ↑ "LSD1 is a subunit of the NuRD complex and targets the metastasis programs in breast cancer". Cell 138 (4): 660–72. Aug 2009. doi:10.1016/j.cell.2009.05.050. PMID 19703393.
- ↑ "KDM1B is a histone H3K4 demethylase required to establish maternal genomic imprints". Nature 461 (7262): 415–8. Sep 2009. doi:10.1038/nature08315. PMID 19727073. Bibcode: 2009Natur.461..415C.
- ↑ "Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3". Molecular Cell 26 (1): 103–15. Apr 2007. doi:10.1016/j.molcel.2007.02.025. PMID 17434130.
- ↑ "Mutation of Drosophila Lsd1 disrupts H3-K4 methylation, resulting in tissue-specific defects during development". Current Biology 17 (9): 808–12. May 2007. doi:10.1016/j.cub.2007.03.068. PMID 17462898.
- ↑ "CpG islands recruit a histone H3 lysine 36 demethylase". Molecular Cell 38 (2): 179–90. Apr 2010. doi:10.1016/j.molcel.2010.04.009. PMID 20417597.
- ↑ "The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b)". Nature Structural & Molecular Biology 15 (11): 1169–75. Nov 2008. doi:10.1038/nsmb.1499. PMID 18836456.
- ↑ "Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells". Genes & Development 21 (20): 2545–57. Oct 2007. doi:10.1101/gad.1588207. PMID 17938240.
- ↑ "Comprehensive genomic analysis of desmoplastic medulloblastomas: identification of novel amplified genes and separate evaluation of the different histological components". The Journal of Pathology 208 (4): 554–63. Mar 2006. doi:10.1002/path.1925. PMID 16400626.
- ↑ "Genomic amplification and oncogenic properties of the GASC1 histone demethylase gene in breast cancer". Oncogene 28 (50): 4491–500. Dec 2009. doi:10.1038/onc.2009.297. PMID 19784073.
- ↑ "Multiple recurrent genetic events converge on control of histone lysine methylation in medulloblastoma". Nature Genetics 41 (4): 465–72. Apr 2009. doi:10.1038/ng.336. PMID 19270706.
- ↑ "Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines". Cancer Research 60 (17): 4735–9. Sep 2000. PMID 10987278.
- ↑ "The putative oncogene GASC1 demethylates tri- and dimethylated lysine 9 on histone H3". Nature 442 (7100): 307–11. Jul 2006. doi:10.1038/nature04837. PMID 16732293. Bibcode: 2006Natur.442..307C.
- ↑ "The H3K4 demethylase lid associates with and inhibits histone deacetylase Rpd3". Molecular and Cellular Biology 29 (6): 1401–10. Mar 2009. doi:10.1128/MCB.01643-08. PMID 19114561.
- ↑ "Binding of pRB to the PHD protein RBP2 promotes cellular differentiation". Molecular Cell 18 (6): 623–35. Jun 2005. doi:10.1016/j.molcel.2005.05.012. PMID 15949438.
- ↑ "Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation". Molecular Cell 31 (4): 520–30. Aug 2008. doi:10.1016/j.molcel.2008.08.004. PMID 18722178.
- ↑ "Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2". Genes & Development 22 (10): 1345–55. May 2008. doi:10.1101/gad.470008. PMID 18483221.
- ↑ "A role for mammalian Sin3 in permanent gene silencing". Molecular Cell 32 (3): 359–70. Nov 2008. doi:10.1016/j.molcel.2008.10.015. PMID 18995834.
- ↑ "The histone demethylase RBP2 Is overexpressed in gastric cancer and its inhibition triggers senescence of cancer cells". Gastroenterology 138 (3): 981–92. Mar 2010. doi:10.1053/j.gastro.2009.10.004. PMID 19850045.
- ↑ "Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation". American Journal of Human Genetics 76 (2): 227–36. Feb 2005. doi:10.1086/427563. PMID 15586325.
- ↑ "The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases". Cell 128 (6): 1077–88. Mar 2007. doi:10.1016/j.cell.2007.02.017. PMID 17320160.
- ↑ "The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence". Genes & Development 23 (10): 1171–6. May 2009. doi:10.1101/gad.510809. PMID 19451217.
- ↑ "Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome". American Journal of Human Genetics 90 (1): 119–24. Jan 2012. doi:10.1016/j.ajhg.2011.11.021. PMID 22197486.
- ↑ "KDM6A point mutations cause Kabuki syndrome". Human Mutation 34 (1): 108–10. Jan 2013. doi:10.1002/humu.22229. PMID 23076834.
- ↑ "A histone H3 lysine 27 demethylase regulates animal posterior development". Nature 449 (7163): 689–94. Oct 2007. doi:10.1038/nature06192. PMID 17851529. Bibcode: 2007Natur.449..689L.
- ↑ 35.0 35.1 "UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development". Nature 449 (7163): 731–4. Oct 2007. doi:10.1038/nature06145. PMID 17713478. Bibcode: 2007Natur.449..731A.
- ↑ "The histone demethylase UTX enables RB-dependent cell fate control". Genes & Development 24 (4): 327–32. Feb 2010. doi:10.1101/gad.1882610. PMID 20123895.
- ↑ "The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing". Cell 130 (6): 1083–94. Sep 2007. doi:10.1016/j.cell.2007.08.019. PMID 17825402.
- ↑ "The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment". PLOS ONE 3 (8): e3034. 2008. doi:10.1371/journal.pone.0003034. PMID 18716661. Bibcode: 2008PLoSO...3.3034B.
- ↑ 39.0 39.1 "Dependence of bacterial chemotaxis on gradient shape and adaptation rate". PLOS Computational Biology 4 (12): e1000242. Dec 2008. doi:10.1371/journal.pcbi.1000242. PMID 19096502. Bibcode: 2008PLSCB...4E0242V.
- ↑ "Reconstruction of the chemotaxis receptor-kinase assembly". Nature Structural & Molecular Biology 13 (5): 400–7. May 2006. doi:10.1038/nsmb1085. PMID 16622408.
- ↑ "Crystal structure of the catalytic domain of the chemotaxis receptor methylesterase, CheB". Journal of Molecular Biology 250 (2): 276–90. Jul 1995. doi:10.1006/jmbi.1995.0376. PMID 7608974.
 |
