Biology:Glycoside hydrolase family 67
| Glycosyl hydrolase family 67 N-terminus | |||||||||
|---|---|---|---|---|---|---|---|---|---|
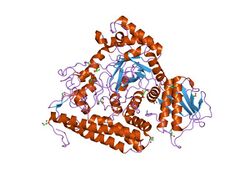 the 1.7 a crystal structure of alpha-d-glucuronidase, a family-67 glycoside hydrolase from bacillus stearothermophilus t-1 | |||||||||
| Identifiers | |||||||||
| Symbol | Glyco_hydro_67N | ||||||||
| Pfam | PF03648 | ||||||||
| InterPro | IPR005154 | ||||||||
| SCOP2 | 1h41 / SCOPe / SUPFAM | ||||||||
| CAZy | GH67 | ||||||||
| |||||||||
| Glycosyl hydrolase family 67 middle domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
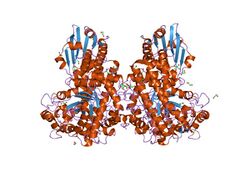 pseudomonas cellulosa e292a alpha-d-glucuronidase mutant complexed with aldotriuronic acid | |||||||||
| Identifiers | |||||||||
| Symbol | Glyco_hydro_67M | ||||||||
| Pfam | PF07488 | ||||||||
| InterPro | IPR011100 | ||||||||
| SCOP2 | 1h41 / SCOPe / SUPFAM | ||||||||
| CAZy | GH67 | ||||||||
| |||||||||
| Glycosyl hydrolase family 67 C-terminus | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 the 1.7 a crystal structure of alpha-d-glucuronidase, a family-67 glycoside hydrolase from bacillus stearothermophilus t-1 | |||||||||
| Identifiers | |||||||||
| Symbol | Glyco_hydro_67C | ||||||||
| Pfam | PF07477 | ||||||||
| InterPro | IPR011099 | ||||||||
| SCOP2 | 1h41 / SCOPe / SUPFAM | ||||||||
| CAZy | GH67 | ||||||||
| |||||||||
In molecular biology, glycoside hydrolase family 67 is a family of glycoside hydrolases.
Glycoside hydrolases EC 3.2.1. are a widespread group of enzymes that hydrolyse the glycosidic bond between two or more carbohydrates, or between a carbohydrate and a non-carbohydrate moiety. A classification system for glycoside hydrolases, based on sequence similarity, has led to the definition of >100 different families.[1][2][3] This classification is available on the CAZy web site,[4][5] and also discussed at CAZypedia, an online encyclopedia of carbohydrate active enzymes.[6][7]
Glycoside hydrolase family 67 includes alpha-glucuronidases, these are components of an ensemble of enzymes central to the recycling of photosynthetic biomass, remove the alpha-1,2 linked 4-O-methyl glucuronic acid from xylans.
Members of this family consist of three structural domains. Deletion mutants of alpha-glucuronidase from Bacillus stearothermophilus have indicated that the central region is responsible for the catalytic activity. Within this central domain, the invariant Glu and Asp (residues 391 and 364 respectively from Bacillus stearothermophilus) are thought to form the catalytic centre.[8] The C-terminal region of alpha-glucuronidase is mainly alpha-helical. It wraps around the catalytic domain, making additional interactions both with the N-terminal domain of its parent monomer and also forming the majority of the dimer-surface with the equivalent C-terminal domain of the other monomer of the dimer.[9]
References
- ↑ "Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases". Proceedings of the National Academy of Sciences of the United States of America 92 (15): 7090–4. July 1995. doi:10.1073/pnas.92.15.7090. PMID 7624375. Bibcode: 1995PNAS...92.7090H.
- ↑ "Structures and mechanisms of glycosyl hydrolases". Structure 3 (9): 853–9. September 1995. doi:10.1016/S0969-2126(01)00220-9. PMID 8535779.
- ↑ "Updating the sequence-based classification of glycosyl hydrolases". The Biochemical Journal 316 ( Pt 2) (Pt 2): 695–6. June 1996. doi:10.1042/bj3160695. PMID 8687420.
- ↑ "Home" (in en). http://www.cazy.org/.
- ↑ "The carbohydrate-active enzymes database (CAZy) in 2013". Nucleic Acids Research 42 (Database issue): D490–5. January 2014. doi:10.1093/nar/gkt1178. PMID 24270786.
- ↑ "Glycoside Hydrolase Family 67" (in en). http://www.cazypedia.org/index.php/Glycoside_Hydrolase_Family_67.
- ↑ CAZypedia Consortium (December 2018). "Ten years of CAZypedia: a living encyclopedia of carbohydrate-active enzymes". Glycobiology 28 (1): 3–8. doi:10.1093/glycob/cwx089. PMID 29040563. https://hal.archives-ouvertes.fr/hal-01886461/file/Hehemann_2018_01.pdf.
- ↑ "Biochemical characterization and identification of catalytic residues in alpha-glucuronidase from Bacillus stearothermophilus T-6". Eur. J. Biochem. 268 (10): 3006–3016. 2001. doi:10.1046/j.1432-1327.2001.02193.x. PMID 11358519.
- ↑ "The structural basis for catalysis and specificity of the Pseudomonas cellulosa alpha-glucuronidase, GlcA67A". Structure 10 (4): 547–56. April 2002. doi:10.1016/s0969-2126(02)00742-6. PMID 11937059.
 |

