Biology:Polygalacturonase
| Endo-polygalacturonase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
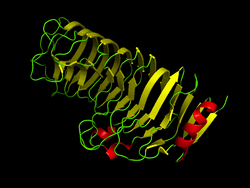 Computer generated image of Polygalacturonase as found in Aspergillus aculeatus (1IA5) at pH 8.5[1] | |||||||||
| Identifiers | |||||||||
| EC number | 3.2.1.15 | ||||||||
| CAS number | 9032-75-1 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Endo-polygalacturonase (EC 3.2.1.15, pectin depolymerase, pectolase, pectin hydrolase, and poly-α-1,4-galacturonide glycanohydrolase; systematic name (1→4)-α-D-galacturonan glycanohydrolase (endo-cleaving)) is an enzyme that hydrolyzes the α-1,4 glycosidic bonds between galacturonic acid residues:
- (1,4-α-D-galacturonosyl)n+m + H2O = (1,4-α-D-galacturonosyl)n + (1,4-α-D-galacturonosyl)m
Polygalacturonan, whose major component is galacturonic acid,[2] is a significant carbohydrate component of the pectin network that comprises plant cell walls.[3] Therefore, the activity of the endogenous plant PGs works to soften and sweeten fruit during the ripening process. Similarly, phytopathogens use PGs as a means to weaken the pectin network, so that digestive enzymes can be excreted into the plant host to acquire nutrients.
Structure
This enzyme's multiple parallel β sheets form a helical shape that is called a β helix. This highly stable structure, thanks to numerous hydrogen bonds and disulfide bonds between strands, is a common characteristic of enzymes involved in the degradation of pectin.[4] The interior of the β helix is hydrophobic.[1]

X-ray crystallography has been used to determine the three-dimensional structure of several PGs in different organisms. Fungal PGs from Colletotrichum lupini,[6] Aspergillus aculeatus,[1] and Aspergillus niger (PG1[7] and PG2[8]) have been crystallized. The PGs from bacteria like Erwinia carotovora[9] and Bacillus subtilis[4] have also been crystallized.
The active site of Fusarium moniliforme PG comprises six charged amino acid residues: H188, R267, and K269 are involved in substrate binding, D212 (a general acid) is responsible for proton donation to the glycosydic oxygen, and D213 and D191 activate H2O for a nucleophilic attack.[5]
Mechanism
Polygalacturonase is a pectinase, an enzyme that degrades pectin by hydrolyzing the O-glycosyl bonds in pectin's polygalacturonan network, resulting in α-1,4-polygalacturonic residues.[10] The rate of hydrolysis is dependent on polysaccharide chain length. Low rates of hydrolysis are associated with very short chains (e.g. digalacturonic acid) and very long chains.[11]

Exo- vs Endo-polygalacturonases
Exo- and Endo-polygalacturonases utilize different hydrolytic modes of action. Endo-polygalacturonases hydrolyze in a random fashion along the polygalacturonan network. This method results in oligogalacturonides.[12] Exo-polygalacturonases hydrolyze at the non-reducing end of the polymer, generating a monosaccharide galacturonic acid. Occasionally, organisms employ both methods. In addition to different modes of action, polygalacturonases polymorphism allows fungal polygalacturonases to more effectively degrade a wider range of plant tissues. PG variety in optimal pH, substrate specificity, and other factors are likely helpful for phytopathogenic organisms like fungi.[4]
Agricultural relevance
Due to the applicability of this enzyme's activity on agricultural productivity and commercial success, much of the research on PGs has revolved around the role of PGs in the fruit ripening process, pollen, and abscission.
Pectin is one of the three polysaccharides present in the plant cell wall, and it plays a role in maintaining the barrier between the inside and outside environment and gives strength to the plant cell walls.[13] Specifically, pectin in the middle lamella holds neighboring cells together.[14]
Fruit ripening
The first GM food available in stores was a genetically modified tomato (also known as Flavr Savr) that had a longer shelf life and was ideal for shipping. Its delayed ripening was achieved by preventing polygalacturonase from destroying pectin, which makes tomatoes firm. An antisense PG gene was introduced, preventing polygalacturonase from ripening and softening the tomato.[15] Although this method has been shown to reduce PG enzymatic activity by 70 to 90%, the PG antisense RNA did not hinder normal color development.[16]
Depolymerization of pectin is largely involved in the later stages of fruit ripening, especially as the fruit becomes overripe.[17] While tomatoes are the prime example of high PG activity, this enzyme is also very active in avocado and peach ripening. PG enzymes in peach, two exo-PGs and one endo-PG, become active when the fruit is already soft.[18] Fruits like persimmons may either lack PG enzymes or have very low levels of PG and as such they have not been detected yet. In these cases, other enzymes may catalyze the ripening process.
Pollen
Exo-PGs play a role in enabling pollen tube elongation since pectin rearrangement is necessary for the growth of pollen tubes. This PG activity has been found in grasses like maize as well as in trees, particularly in the Eastern cottonwood.[19] Exo-PGs involved in pollen tube growth need Ca2+ for maximal enzymatic activity and can be inhibited by high concentrations of NaCl, citrate, and EDTA.[11]
Abscission zones
It is largely unclear whether PGs play a role in facilitating abscission in certain plants, and if they do, whether they are exo- or endo-acting. Conflicting research has been published on, for example, whether PG is involved in citrus fruit abscission. One particular issue has been the usage of assays that are not able to measure exo-PG activity.[20] An additional complication is the difference in PG enzymatic activity between fruit and leaf cell-separation zones. In peach, PG activity was only detected in fruit abscission zones.[21]
Other
Agricultural pests like Lygus hesperus damage cotton and other crops because they secrete PGs in their saliva that digest plant tissue. They employ both exo- and endo-PGs.[22]
Inhibition

Phytopathogenic fungi expose plant cell walls to cell wall degrading enzymes (CWDEs) like PGs.[4] In response, most plants have natural inhibitor proteins that slow the hydrolytic activity of PG. These inhibitors also prompt long chain oligogalacturonide accumulation in order to encourage a defense mechanism against the attack.[4] The polygalacturonase inhibitor proteins (PGIPs) are leucine-rich repeat proteins that have been reported to demonstrate both non-competitive[24] and competitive[5] inhibition of PGs. The active site of PG interacts with a pocket containing multiple polar amino acids in Phaseolus vulgaris PGIP2. The inhibitor prevents substrate binding by occupying the active site, resulting in competitive inhibition.[23]
The crystal structures for PGIP and PGIP2 have been determined for the bean P. vulgaris. The charged and polar residues that interact with the PG active site have been identified in P. vulgaris as D131, S133, T155, D157, T180, and D203.[23] Using PGIP2 as a template, the theoretical structures of other PGIPs have been determined for some other common crops.
References
- ↑ 1.0 1.1 1.2 "The X-ray structure of Aspergillus aculeatus polygalacturonase and a modeled structure of the polygalacturonase-octagalacturonate complex". Journal of Molecular Biology 311 (4): 863–78. August 2001. doi:10.1006/jmbi.2001.4919. PMID 11518536.
- ↑ "Medical Definition of Polygalacturonan". http://www.lexic.us/definition-of/polygalacturonan.
- ↑ Jones, T. M., Anderson, A. J., and Albersheim, P. (1972) Host–pathogen interactions IV, Studies on the polysaccharide-degrading enzymes secreted by Fusarium oxysporum f. sp. lycopersici, Physiol. Plant Pathol. 2, 153-166.
- ↑ 4.0 4.1 4.2 4.3 4.4 "Polygalacturonases, polygalacturonase-inhibiting proteins and pectic oligomers in plant-pathogen interactions". Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1696 (2): 237–44. February 2004. doi:10.1016/j.bbapap.2003.08.012. PMID 14871664.
- ↑ 5.0 5.1 5.2 Federici L, Caprari C, Mattei B, Savino C, Di Matteo A, De Lorenzo G, Cervone F, Tsernoglou D. Structural requirements of endopolygalacturonase for the interaction with PGIP (polygalacturonase-inhibiting protein). Proc Natl Acad Sci U S A. 2001 Nov 6;98(23):13425-30.
- ↑ "Crystal structure of the endopolygalacturonase from the phytopathogenic fungus Colletotrichum lupini and its interaction with polygalacturonase-inhibiting proteins". Proteins 70 (1): 294–9. January 2008. doi:10.1002/prot.21610. PMID 17876815.
- ↑ "Structural insights into the processivity of endopolygalacturonase I from Aspergillus niger". FEBS Letters 554 (3): 462–6. November 2003. doi:10.1016/s0014-5793(03)01221-3. PMID 14623112. https://pure.rug.nl/ws/files/6675169/2003FEBSLettvPouderoyen.pdf.
- ↑ "1.68-A crystal structure of endopolygalacturonase II from Aspergillus niger and identification of active site residues by site-directed mutagenesis". The Journal of Biological Chemistry 274 (43): 30474–80. October 1999. doi:10.1074/jbc.274.43.30474. PMID 10521427.
- ↑ "Crystal structure of polygalacturonase from Erwinia carotovora ssp. carotovora". The Journal of Biological Chemistry 273 (38): 24660–4. September 1998. doi:10.1074/jbc.273.38.24660. PMID 9733763.
- ↑ 10.0 10.1 "Information on EC 3.2.1.15 - polygalacturonase". http://www.brenda-enzymes.org/enzyme.php?ecno=3.2.1.15.
- ↑ 11.0 11.1 Pressey, Russell; Reger, Bonnie J. (1989). "Polygalacturonase in pollen from corn and other grasses". Plant Science 59 (1): 57–62. doi:10.1016/0168-9452(89)90008-3.
- ↑ "Oligogalacturonides: plant damage-associated molecular patterns and regulators of growth and development". Frontiers in Plant Science 4 (49): 49. 13 Mar 2013. doi:10.3389/fpls.2013.00049. PMID 23493833.
- ↑ "Biosynthesis of pectin". Plant Physiology 153 (2): 384–95. June 2010. doi:10.1104/pp.110.156588. PMID 20427466.
- ↑ "Altered middle lamella homogalacturonan and disrupted deposition of (1→5)-α-L-arabinan in the pericarp of Cnr, a ripening mutant of tomato". Plant Physiology 126 (1): 210–21. May 2001. doi:10.1104/pp.126.1.210. PMID 11351084.
- ↑ Berg, Jeremy M.; Tymoczko, John L.; Stryer, Lubert (2012). Biochemistry (7 ed.). W. H. Freeman and Company. p. 167. ISBN 9781429229364.
- ↑ "Reduction of polygalacturonase activity in tomato fruit by antisense RNA". Proceedings of the National Academy of Sciences of the United States of America 85 (23): 8805–9. December 1988. doi:10.1073/pnas.85.23.8805. PMID 16593997. Bibcode: 1988PNAS...85.8805S.
- ↑ "Polygalacturonases: many genes in search of a function". Plant Physiology 117 (2): 337–43. June 1998. doi:10.1104/pp.117.2.337. PMID 9625687.
- ↑ G. Downs, Christopher; J. Brady, Colin; Gooley, Andrew (June 1992). "Exopolygalacturonase protein accumulates late in peach fruit ripening". Physiologia Plantarum 85 (2): 133–140. doi:10.1111/j.1399-3054.1992.tb04715.x.
- ↑ Pressey, Russell (1991). "Polygalacturonase in tree pollens". Phytochemistry 30 (6): 1753–1755. doi:10.1016/0031-9422(91)85006-L. https://zenodo.org/record/1258252.
- ↑ Berger, Richard K.; Reid, Philip D. (15 January 1979). "Role of Polygalacturonase in Bean Leaf Abscission". Plant Physiology 63 (6): 1133–1137. doi:10.1104/pp.63.6.1133. PMID 16660870.
- ↑ Bonghi, C.; Rascio, N.; Ramina, A.; Casadoro, G. (December 1992). "Cellulase and polygalacturonase involvement in the abscission of leaf and fruit explants of peach". Plant Molecular Biology 20 (5): 839–848. doi:10.1007/bf00027155. PMID 1281437.
- ↑ "Identification of endo- and exo-polygalacturonase activity in Lygus hesperus (Knight) salivary glands". Archives of Insect Biochemistry and Physiology 70 (2): 122–35. February 2009. doi:10.1002/arch.20282. PMID 19085947. http://www.plantsciences.ucdavis.edu/faculty/labavitch/PDFs/Celorio-Mancera%20et%20al.2009.pdf.
- ↑ 23.0 23.1 23.2 "The crystal structure of polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein involved in plant defense". Proceedings of the National Academy of Sciences of the United States of America 100 (17): 10124–8. August 2003. doi:10.1073/pnas.1733690100. PMID 12904578. Bibcode: 2003PNAS..10010124D.
- ↑ Daniel King, Carl Bergmann, Ron Orlando, Jacques A. E. Benen, Harry C. M. Kester, and Jaap Visser; "Use of Amide Exchange Mass Spectrometry To Study Conformational Changes within the Endopolygalacturonase II – Polygalacturonic Acid – Polygalacturonase Inhibiting Protein System", Biochem. 41, 10225-10233, 2002
External links
 |
