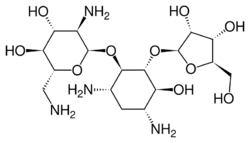
Chemistry:Ribostamycin
 | |
| Clinical data | |
|---|---|
| Other names | (2R,3S,4R,5R,6R)-5-amino-2-(aminomethyl)-6-{[(1R,2R,3S,4R,6S)-4,6-diamino-2-{[(2S,3R,4S,5R)-3,4-dihydroxy-5-(hydroxymethyl)oxolan-2-yl]oxy}-3-hydroxycyclohexyl]oxy}oxane-3,4-diol |
| AHFS/Drugs.com | International Drug Names |
| ATC code | |
| Identifiers | |
| |
| CAS Number |
|
| PubChem CID | |
| ChemSpider | |
| UNII | |
| ChEMBL | |
| Chemical and physical data | |
| Formula | C17H34N4O10 |
| Molar mass | 454.477 g·mol−1 |
| 3D model (JSmol) | |
| |
| |
| | |
Ribostamycin is an aminoglycoside-aminocyclitol antibiotic isolated from a streptomycete, Streptomyces ribosidificus, originally identified in a soil sample from Tsu City of Mie Prefecture in Japan.[1] It is made up of 3 ring subunits: 2-deoxystreptamine (DOS), neosamine C, and ribose.[2] Ribostamycin, along with other aminoglycosides with the DOS subunit, is an important broad-spectrum antibiotic with important use against human immunodeficiency virus[citation needed] and is considered a critically important antimicrobial by the World Health Organization.,[3][4] Resistance against aminoglycoside antibiotics, such as ribostamycin, is a growing concern. The resistant bacteria contain enzymes that modify the structure through phosphorylation, adenylation, and acetylation and prevent the antibiotic from being able to interact with the bacterial ribosomal RNAs.[5]
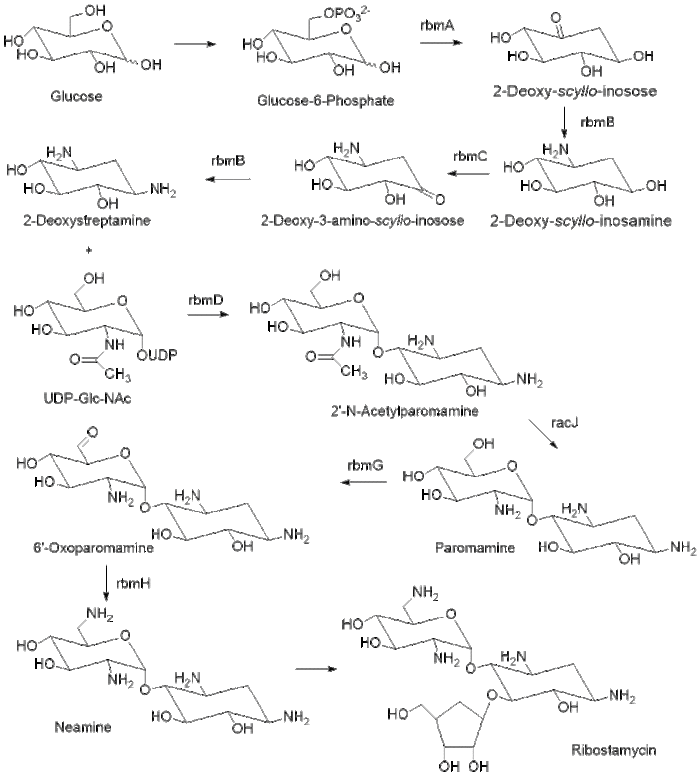
Biosynthesis
The biosynthesis of ribostamycin begins with the sugar, D-glucose, which is phosphorylated at the 6 position to form glucose-6-phosphate. The enzyme rbmA contains a genetic sequence that corresponds to NAD+ binding and catalyzes the formation of 2-deoxy-scyllo-inosose. The enzyme rmbB then catalyzes the transamination of 2-deoxy-scyllo-inosose to 2-deoxy-scyllo-inosamine with L-glutamine and pyridoxal phosphate (PLP). Enzyme rbmC oxidizes the ring to 2-deoxy-3-amino-scyllo-inosose, which is then transaminated by enzyme rmbB to DOS. DOS is then glycosylated by the glycosyltransferase rmbD with uridine diphosphate N-acetylglucosamine (UDP-Glc-NAc) to form 2’-N-acetylparomamine. The deacetylase, racJ, removes the acetyl group and forms paromamine. Paromamine is oxidized by enzyme rbmG and then enzyme rmbH transaminates to produce neamine. Neamine is then ribosylated to form ribostamycin.,[2][3]
References
- ↑ "Studies on antibiotic SF-733, a new antibiotic. I. Taxonomy, isolation and characterization". The Journal of Antibiotics 23 (3): 155–61. March 1970. doi:10.7164/antibiotics.23.155. PMID 5453309.
- ↑ 2.0 2.1 "The ribostamycin biosynthetic gene cluster in Streptomyces ribosidificus: comparison with butirosin biosynthesis". Molecules and Cells 20 (1): 90–6. August 2005. PMID 16258246.
- ↑ 3.0 3.1 "Biosynthesis of ribostamycin derivatives by reconstitution and heterologous expression of required gene sets". Applied Biochemistry and Biotechnology 163 (3): 373–82. February 2011. doi:10.1007/s12010-010-9045-6. PMID 20676801.
- ↑ WHO Advisory Group on Integrated Surveillance of Antimicrobial Resistance (AGISAR) (2011). Critically Important Antimicrobials for Human Medicine (Report) (3rd revision ed.). World Health Organization.. ISBN 978-92-4-150448-5. http://apps.who.int/iris/bitstream/10665/77376/1/9789241504485_eng.pdf.
- ↑ "Biosynthetic genes for aminoglycoside antibiotics". The Journal of Antibiotics 62 (9): 471–81. September 2009. doi:10.1038/ja.2009.76. PMID 19644520.
 |