Biology:Hepacivirus
| Hepacivirus | |
|---|---|
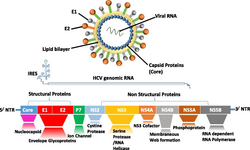
| |
| Hepacivirus structure and genome | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Kitrinoviricota |
| Class: | Flasuviricetes |
| Order: | Amarillovirales |
| Family: | Flaviviridae |
| Genus: | Hepacivirus |
| Synonyms | |
Hepacivirus is a genus of positive-strand RNA viruses in the family Flaviviridae.[3] The hepatitis C virus (HCV), in species Hepacivirus C, infects humans and is associated with hepatitis and hepatocellular carcinoma.[3][4] There are fourteen species in the genus which infect a range of other vertebrate.[3]
History
Hepatitis C virus (HCV), which is the causative agent of hepatitis C in humans, and a member of the species Hepacivirus C, was discovered in 1989.[5] Seven genotypes (1–7) and eighty-six subtypes (1a, 1b etc.) of hepatitis C virus have been named.[6]
GBV-B virus (also known as GB virus B) discovered in 1995 is capable of infecting New World monkeys, in particular tamarins. Like HCV it is transmitted by the blood-borne route and similar to HCV it is associated with the viral hepatitis. However GBV-B has never been identified in wild animals and its natural host is not known.[5]
Structure
Viruses in the genus Hepacivirus are enveloped and have spherical icosahedral-like geometries with pseudo T=3 symmetry. The virus particle diameter is around 50 nm. Genomes are linear, non-segmented, and around 10,000 nucleotides in length.[3][4]
Life cycle
Entry into the host cell is achieved by attachment of the viral envelope protein E to host receptors, which mediates clathrin-mediated endocytosis. Replication follows the positive-strand RNA virus replication model. Positive strand RNA virus transcription is the method of transcription. Translation takes place by viral initiation. Humans and other vertebrate serve as the natural host. Transmission routes are sexual, blood, and contact.[3][4]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Hepacivirus | Humans | Epithelium: skin; epithelium: kidney; epithelium: intestine; epithelium: testes | Clathrin-mediated endocytosis | Secretion | Cytoplasm | Cytoplasm | Sex; blood |
Taxonomy

In the genus Hepacvirus there are 14 species:[7][3]
Additional information
Additional hepaciviruses have been described from bats, rodents including bank voles, horses, and dogs.[8][9][10] Rodent hepacivirus is found in the deer mouse (Peromyscus maniculatus).[5] A virus related to hepaciviruses is found in bamboo rat (Rhizomys pruinosus).[11] A virus related to hepaciviruses infects the long-tailed ground squirrel Spermophilus undulatus.[12]
Cattle are a host for viruses of the species Hepacivirus N.[13][14] The viruses most closely related to Hepacivirus C are the equine hepaciviruses of the species Hepacivirus A.[15] There are at least two subtypes of equine hepacivirus.[16] Hepacivirus A infecting horses has also been found in donkeys.[17]
A virus related to the hepaciviruses has been isolated from bald eagles (Haliaeetus leucocephalus).[18] Another unclassified virus in this taxon is duck hepacivirus-like virus.[19] A virus related to hepaciviruses has been isolated from the graceful catshark (Proscyllium habereri).[20] The virus – Jogalong virus – has been described that appears to belong to another species in this genus.[21]
References
- ↑ Pringle, C. R. (1996). "Virus Taxonomy 1996 - A Bulletin from the Xth International Congress of Virology in Jerusalem" (in en). Archives of Virology 141 (11): 2251–6. doi:10.1007/BF01718231. ISSN 1432-8798. OCLC 76349762. PMID 8992952. PMC 7086844. https://ictv.global/ictv/proposals/Ratification_1996.pdf. Retrieved 13 March 2019. "To name the genus comprising the hepatitis C-like viruses as the genus Hepacivirus.".
- ↑ 1ICTV 6th Report Murphy, F. A., Fauquet, C. M., Bishop, D. H. L., Ghabrial, S. A., Jarvis, A. W. Martelli, G. P. Mayo, M. A. & Summers, M. D.(eds)(1995). Virus Taxonomy. Sixthreport of the International Committee on Taxonomy of Viruses. Archives of Virology Supplement 10, p. 424 https://ictv.global/ictv/proposals/ICTV%206th%20Report.pdf
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 "Flaviviridae" (in en). http://www.ictv.global/report/flaviviridae.
- ↑ 4.0 4.1 4.2 "Viral Zone". ExPASy. http://viralzone.expasy.org/all_by_species/37.html. Retrieved 15 June 2015.
- ↑ 5.0 5.1 5.2 Stapleton, J. T; Foung, S; Muerhoff, A. S; Bukh, J; Simmonds, P (2010). "The GB viruses: A review and proposed classification of GBV-A, GBV-C (HGV), and GBV-D in genus Pegivirus within the family Flaviviridae". Journal of General Virology 92 (2): 233–46. doi:10.1099/vir.0.027490-0. PMID 21084497.
- ↑ "HCV Classification" (in en). http://talk.ictvonline.org/links/hcv/hcv-classification.htm.[|permanent dead link|dead link}}]
- ↑ Smith, Donald B; Becher, Paul; Bukh, Jens; Gould, Ernest A; Meyers, Gregor; Monath, Thomas; Muerhoff, A Scott; Pletnev, Alexander et al. (2016). "Proposed update to the taxonomy of the genera Hepacivirus and Pegivirus within the Flaviviridae family". Journal of General Virology 97 (11): 2894–2907. doi:10.1099/jgv.0.000612. PMID 27692039.
- ↑ Kapoor, A; Simmonds, P; Scheel, T. K. H; Hjelle, B; Cullen, J. M; Burbelo, P. D; Chauhan, L. V; Duraisamy, R et al. (2013). "Identification of Rodent Homologs of Hepatitis C Virus and Pegiviruses". mBio 4 (2): e00216–13. doi:10.1128/mBio.00216-13. PMID 23572554.
- ↑ Drexler, Jan Felix; Corman, Victor Max; Müller, Marcel Alexander; Lukashev, Alexander N; Gmyl, Anatoly; Coutard, Bruno; Adam, Alexander; Ritz, Daniel et al. (2013). "Evidence for Novel Hepaciviruses in Rodents". PLOS Pathogens 9 (6): e1003438. doi:10.1371/journal.ppat.1003438. PMID 23818848.
- ↑ Lauck, M; Sibley, S. D; Lara, J; Purdy, M. A; Khudyakov, Y; Hyeroba, D; Tumukunde, A; Weny, G et al. (2013). "A Novel Hepacivirus with an Unusually Long and Intrinsically Disordered NS5A Protein in a Wild Old World Primate". Journal of Virology 87 (16): 8971–81. doi:10.1128/JVI.00888-13. PMID 23740998.
- ↑ Van Nguyen, Dung; Van Nguyen, Cuong; Bonsall, David; Ngo, Tue; Carrique-Mas, Juan; Pham, Anh; Bryant, Juliet; Thwaites, Guy et al. (2018). "Detection and Characterization of Homologues of Human Hepatitis Viruses and Pegiviruses in Rodents and Bats in Vietnam". Viruses 10 (3): 102. doi:10.3390/v10030102. PMID 29495551.
- ↑ Li LL, Liu MM, Shen S, Zhang YJ, Xu YL, Deng HY, Deng F, Duan ZJ (2019) Detection and characterization of a novel hepacivirus in long-tailed ground squirrels (Spermophilus undulatus) in China. Arch Virol
- ↑ Corman, Victor Max; Grundhoff, Adam; Baechlein, Christine; Fischer, Nicole; Gmyl, Anatoly; Wollny, Robert; Dei, Dickson; Ritz, Daniel et al. (2015). "Highly Divergent Hepaciviruses from African Cattle". Journal of Virology 89 (11): 5876–82. doi:10.1128/JVI.00393-15. PMID 25787289.
- ↑ Baechlein, Christine; Fischer, Nicole; Grundhoff, Adam; Alawi, Malik; Indenbirken, Daniela; Postel, Alexander; Baron, Anna Lena; Offinger, Jennifer et al. (2015). "Identification of a Novel Hepacivirus in Domestic Cattle from Germany". Journal of Virology 89 (14): 7007–15. doi:10.1128/JVI.00534-15. PMID 25926652.
- ↑ Thézé, Julien; Lowes, Sophia; Parker, Joe; Pybus, Oliver G (2015). "Evolutionary and Phylogenetic Analysis of the Hepaciviruses and Pegiviruses". Genome Biology and Evolution 7 (11): 2996–3008. doi:10.1093/gbe/evv202. PMID 26494702.
- ↑ Pronost, S; Hue, E; Fortier, C; Foursin, M; Fortier, G; Desbrosse, F; Rey, F. A; Pitel, P.-H et al. (2017). "Prevalence of Equine Hepacivirus Infections in France and Evidence for Two Viral Subtypes Circulating Worldwide". Transboundary and Emerging Diseases 64 (6): 1884–1897. doi:10.1111/tbed.12587. PMID 27882682.
- ↑ Walter S, Rasche A, Moreira-Soto A, Pfaender S, Bletsa M, Corman VM, Aguilar-Setien A, García-Lacy F, Hans A, Todt D, Schuler G, Shnaiderman-Torban A, Steinman A, Roncoroni C, Veneziano V, Rusenova N12, Sandev N12, Rusenov A, Zapryanova D, García-Bocanegra I, Jores J, Carluccio A, Veronesi MC, Cavalleri JMV, Drosten C, Lemey P, Steinmann E, Drexler JF (2016) Differential infection patterns and recent evolutionary origins of Equine Hepaciviruses in donkeys. J Virol 91(1)
- ↑ Goldberg TL, Sibley SD, Pinkerton ME, Dunn CD, Long LJ, White LC, Strom SM (2019) Multidecade mortality and a homolog of Hepatitis C Virus in bald eagles (Haliaeetus leucocephalus), the national bird of the USA. Sci Rep 9(1):14953
- ↑ Chu L, Jin M, Feng C, Wang X, Zhang D (2019) A highly divergent hepacivirus-like flavivirus in domestic ducks. J Gen Virol
- ↑ Shi, Mang; Lin, Xian-Dan; Vasilakis, Nikos; Tian, Jun-Hua; Li, Ci-Xiu; Chen, Liang-Jun; Eastwood, Gillian; Diao, Xiu-Nian et al. (2016). "Divergent Viruses Discovered in Arthropods and Vertebrates Revise the Evolutionary History of the Flaviviridae and Related Viruses". Journal of Virology 90 (2): 659–69. doi:10.1128/JVI.02036-15. PMID 26491167.
- ↑ Williams SH, Levy A, Yates RA, Somaweera N, Neville PJ, Nicholson J, Lindsay MDA, Mackenzie JS, Jain K, Imrie A, Smith DW, Lipkin WI (2020) Discovery of Jogalong virus, a novel hepacivirus identified in a Culex annulirostris (Skuse) mosquito from the Kimberley region of Western Australia. PLoS One 15(1):e0227114
External links
- ICTV Report: Flaviviridae
- Viralzone: Hepacivirus
- Virus Pathogen Database and Analysis Resource (ViPR): Flaviviridae
Wikidata ☰ Q2082778 entry
 |

