Biology:Crossover junction endodeoxyribonuclease
| Crossover junction endodeoxyribonuclease | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 3.1.22.4 | ||||||||
| CAS number | 99676-43-4 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
Crossover junction endodeoxyribonuclease, also known as Holliday junction resolvase, Holliday junction endonuclease, Holliday junction-cleaving endonuclease, Holliday junction-resolving endoribonuclease, crossover junction endoribonuclease, and cruciform-cutting endonuclease, is an enzyme involved in DNA repair and homologous recombination. Specifically, it performs endonucleolytic cleavage that results in single-stranded crossover between two homologous DNA molecules at the Holliday junction to produce recombinant DNA products for chromosomal segregation. This process is known as Holliday junction resolution.
Biological Function
The Holliday junction is a structure that forms during genetic recombination, and links two double-stranded DNA molecules with a single-stranded crossover, which form during mitotic and meiotic recombination.[1] Crossover junction endodeoxyribonucleases catalyze Holiday junction resolution, which is the formation of separate recombinant DNA molecules and chromosomal separation after the crossover event at the Holliday junction.[2] Crossover junction endodeoxyribonucleases with Holliday Junction resolution function have been identified in all three domains of life - bacteria, archaea, and eukarya. RuvC in bacteria, CCE1 in Saccharomyces cerevisiae,[1] and GEN1 in humans [3] are all crossover junction endodeoxyribonucleases that perform Holliday Junction resolution. Holliday junction resolution catalyzed by crossover junction endodeoxyribonuclease is shown in the figure below.
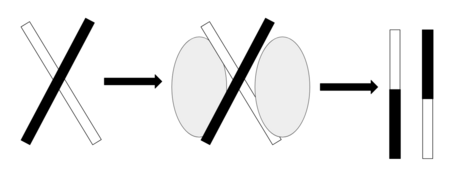
Crossover junction endodeoxyribonucleases also play key roles in DNA repair. During cell growth and meiosis, DNA double-strand breaks (DSBs) often occur, and are usually repaired by homologous recombination.[5] Because Crossover junction endodeoxyribonucleases perform Holliday Junction resolution, a crucial step of homologous recombination, they are therefore involved in repair of DSBs.
Structure
E. coli RuvC, a Crossover junction endodeoxyribonuclease, is a small protein of about 20 kD, and its active form is a dimer that requires and binds a magnesium ion [1]. RuvC is a 3-layer alpha-beta sandwich with a beta-sheet between 5 alpha-helices[6] . The enzyme contains two binding channels that contact the backbones of the Holliday junction over seven nucleotides.[7] A Holliday junction resolvase enzyme has also been identified in archaea in Pyrococcus furiosus cells - it is encoded by a gene called hjc and is composed of 123 amino acids [8] .
A figure of Thermus thermophilus RuvC in complex with a Holliday junction is shown below.
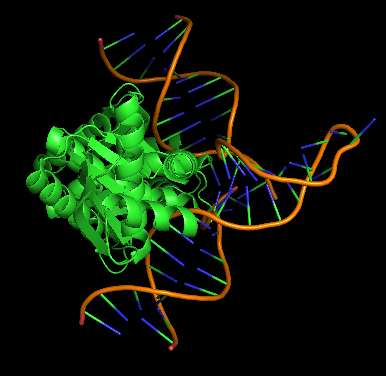
Mechanism
These enzymes are highly selective for branched DNA, although induced fit occurs in the enzyme-substrate (resolvase-Holloday Junction) complex formation.[9] Much remains unknown about the exact mechanism of action, but it is known that bacteria, bacteriophages and archaea catalyze Holliday junction resolution by introducing symmetric nicks across the Holliday junction [10] . Analysis of crossover junction endodeoxyribonucleases from bacteriophages (T7 endonuclease I), bacteria (RuvC), fungi (GEN1) and humans (hMus81-Eme1) have revealed that the enzymes function in dimers,[11] and part of the resolution reaction takes place in a partially dissociated enzyme-substrate intermediate.[12]
Human Relevance
After a 20-year search, in 2008, a human crossover junction endodeoxyribonuclease, GEN1, was finally identified [13] . GEN1 performs similar functions and operates by similar mechanisms as previously studied Crossover junction endodeoxyribonuclease in bacteria, archaea, and other eukarya.[13] The enzyme is thought to play a role in Bloom's syndrome. It has been proposed that Bloom's syndrome involves the induction of DSBs via an unidentified Holliday junction resolvase.[14] It has also been shown that overexpression of Holliday Junction resolvase function is correlated with RAD51-overexpressing cancers.[15]
References
- ↑ 1.0 1.1 "Escherichia coli RuvC protein is an endonuclease that resolves the Holliday structure". The EMBO Journal 10 (13): 4381–9. December 1991. doi:10.1002/j.1460-2075.1991.tb05016.x. PMID 1661673.
- ↑ "Interaction of branch migration translocases with the Holliday junction-resolving enzyme and their implications in Holliday junction resolution". The Journal of Biological Chemistry 289 (25): 17634–46. June 2014. doi:10.1074/jbc.M114.552794. PMID 24770420.
- ↑ "Identification of Holliday junction resolvases from humans and yeast". Nature 456 (7220): 357–61. November 2008. doi:10.1038/nature07470. PMID 19020614. Bibcode: 2008Natur.456..357I.
- ↑ "Holliday junction resolvases". Cold Spring Harbor Perspectives in Biology 6 (9): a023192. September 2014. doi:10.1101/cshperspect.a023192. PMID 25183833.
- ↑ "The role of Holliday junction resolvases in the repair of spontaneous and induced DNA damage". Nucleic Acids Research 39 (16): 7009–19. September 2011. doi:10.1093/nar/gkr277. PMID 21609961.
- ↑ 6.0 6.1 "Crystal structure of RuvC resolvase in complex with Holliday junction substrate". Nucleic Acids Research 41 (21): 9945–55. November 2013. doi:10.1093/nar/gkt769. PMID 23980027.
- ↑ "Holliday junction-resolving enzymes-structures and mechanisms". FEBS Letters 591 (8): 1073–1082. April 2017. doi:10.1002/1873-3468.12529. PMID 27990631. https://discovery.dundee.ac.uk/ws/files/11272660/Lilley_2016_FEBS_Letters_1_.pdf.
- ↑ "A Holliday junction resolvase from Pyrococcus furiosus: functional similarity to Escherichia coli RuvC provides evidence for conserved mechanism of homologous recombination in Bacteria, Eukarya, and Archaea". Proceedings of the National Academy of Sciences of the United States of America 96 (16): 8873–8. August 1999. doi:10.1073/pnas.96.16.8873. PMID 10430863. Bibcode: 1999PNAS...96.8873K.
- ↑ "Mechanism of Holliday junction resolution by the human GEN1 protein". Genes & Development 24 (14): 1559–69. July 2010. doi:10.1101/gad.585310. PMID 20634321.
- ↑ "The structural basis of Holliday junction resolution by T7 endonuclease I". Nature 449 (7162): 621–4. October 2007. doi:10.1038/nature06158. PMID 17873858. Bibcode: 2007Natur.449..621H.
- ↑ "Resolution of single and double Holliday junction recombination intermediates by GEN1". Proceedings of the National Academy of Sciences of the United States of America 114 (3): 443–450. January 2017. doi:10.1073/pnas.1619790114. PMID 28049850.
- ↑ "Junction resolving enzymes use multivalency to keep the Holliday junction dynamic". Nature Chemical Biology 15 (3): 269–275. March 2019. doi:10.1038/s41589-018-0209-y. PMID 30664685.
- ↑ 13.0 13.1 "The search for a human Holliday junction resolvase". Biochemical Society Transactions 37 (Pt 3): 519–26. June 2009. doi:10.1042/BST0370519. PMID 19442245.
- ↑ "The Bloom's syndrome gene product promotes branch migration of holliday junctions". Proceedings of the National Academy of Sciences of the United States of America 97 (12): 6504–8. June 2000. doi:10.1073/pnas.100448097. PMID 10823897. Bibcode: 2000PNAS...97.6504K.
- ↑ "Holliday junction trap shows how cells use recombination and a junction-guardian role of RecQ helicase". Science Advances 2 (11): e1601605. November 2016. doi:10.1126/sciadv.1601605. PMID 28090586. Bibcode: 2016SciA....2E1605X.
External links
- Crossover+junction+endodeoxyribonuclease at the US National Library of Medicine Medical Subject Headings (MeSH)
 |

