Biology:Ubiquitin carboxy-terminal hydrolase L1
 Generic protein structure example |
Ubiquitin carboxy-terminal hydrolase L1 (EC 3.1.2.15, ubiquitin C-terminal hydrolase, UCH-L1) is a deubiquitinating enzyme.
| Ubiquitin Carboxy-terminal Hydrolase L1 (UCH-L1) | |
|---|---|
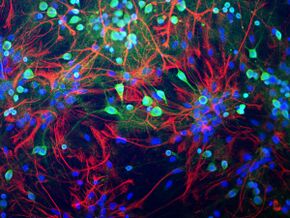 Neurons from rat brain tissue stained green with antibody to ubiquitin C-terminal hydrolase L1 (UCH-L1) which highlights the cell body strongly and the cell processes more weakly. Astrocytes are stained in red with antibody to the GFAP protein found in cytoplasmic filaments. Nuclei of all cell types are stained blue with a DNA binding dye. Antibodies, cell preparation and image generated by EnCor Biotechnology Inc. | |
| Anatomical terms of microanatomy |
Function
UCH-L1 is a member of a gene family whose products hydrolyze small C-terminal adducts of ubiquitin to generate the ubiquitin monomer. Expression of UCH-L1 is highly specific to neurons and to cells of the diffuse neuroendocrine system and their tumors. It is abundantly present in all neurons (accounts for 1-2% of total brain protein), expressed specifically in neurons and testis/ovary.[1][2]
The catalytic triad of UCH-L1 contains a cysteine at position 90, an aspartate at position 176, and a histidine at position 161 that are responsible for its hydrolase activity.[3]
Relevance to neurodegenerative disorders
A point mutation (I93M) in the gene encoding this protein is implicated as the cause of Parkinson's disease in one German family, although this finding is controversial, as no other Parkinson's disease patients with this mutation have been found.[4][5]
Furthermore, a polymorphism (S18Y) in this gene has been found to be associated with a reduced risk for Parkinson's disease.[6] This polymorphism has specifically been shown to have antioxidant activity.[7]
Another potentially protective function of UCH-L1 is its reported ability to stabilize monoubiquitin, an important component of the ubiquitin proteasome system. It is thought that by stabilizing the monomers of ubiquitin and thereby preventing their degradation, UCH-L1 increases the available pool of ubiquitin to be tagged onto proteins destined to be degraded by the proteasome.[8]
The gene is also associated with Alzheimer's disease, and required for normal synaptic and cognitive function.[9] Loss of Uchl1 increases the susceptibility of pancreatic beta-cells to programmed cell death, indicating that this protein plays a protective role in neuroendocrine cells and illustrating a link between diabetes and neurodegenerative diseases.[10]
Patients with early-onset neurodegeneration in which the causative mutation was in the UCHL1 gene (specifically, the ubiquitin binding domain, E7A) display blindness, cerebellar ataxia, nystagmus, dorsal column dysfunction, and upper motor neuron dysfunction.[11]
Ectopic expression
Although UCH-L1 protein expression is specific to neurons and testis/ovary tissue, it has been found to be expressed in certain lung-tumor cell lines.[12] This abnormal expression of UCH-L1 is implicated in cancer and has led to the designation of UCH-L1 as an oncogene.[13] Furthermore there is evidence that UCH-L1 might play a role in the pathogenesis of membranous glomerulonephritis as UCH-L1 de novo expression in podocytes was seen in PHN, the rat model of human mGN.[14] This UCH-L1 expression is thought to induce at least in part podocyte hypertrophy.[15]
Protein structure
Human UCH-L1 and the closely related protein UCHL3 have one of the most complicated knot structure yet discovered for a protein, with five knot crossings. It is speculated that a knot structure may increase a protein's resistance to degradation in the proteasome.[16][17]
The conformation of the UCH-L1 protein may also be an important indication of neuroprotection or pathology. For example, the UCH-L1 dimer has been shown to exhibit the potentially pathogenic ligase activity and may lead to the aforementioned increase in aggregation of α-synuclein.[18] The S18Y polymorphism of UCH-L1 has been shown to be less-prone to dimerization.[8]
Interactions
Ubiquitin carboxy-terminal hydrolase L1 has been shown to interact with COP9 constitutive photomorphogenic homolog subunit 5.[19]
UCH-L1 has also been shown to interact with α-synuclein, another protein implicated in the pathology of Parkinson disease. This activity is reported to be the result of its ubiquityl ligase activity which may be associated with the I93M pathogenic mutation in the gene.[18]
Most recently, UCH-L1 has been demonstrated to interact with the E3 ligase, parkin. Parkin has been demonstrated to bind and ubiquitinylate UCH-L1 to promote lysosomal degradation of UCH-L1.[20]
See also
- Ubiquitin carboxyl-terminal esterase L3—the gene UCHL3
- Alpha synuclein
- Parkinson disease
- Proteasome
References
- ↑ "Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis". Journal of Neurochemistry 40 (6): 1542–7. Jun 1983. doi:10.1111/j.1471-4159.1983.tb08124.x. PMID 6343558.
- ↑ "Entrez Gene: UCHL1 ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)". https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=ShowDetailView&TermToSearch=7345.
- ↑ "Structural basis for conformational plasticity of the Parkinson's disease-associated ubiquitin hydrolase UCH-L1". Proceedings of the National Academy of Sciences of the United States of America 103 (12): 4675–80. Mar 2006. doi:10.1073/pnas.0510403103. PMID 16537382. Bibcode: 2006PNAS..103.4675D.
- ↑ "The ubiquitin pathway in Parkinson's disease". Nature 395 (6701): 451–2. Oct 1998. doi:10.1038/26652. PMID 9774100. Bibcode: 1998Natur.395..451L.
- ↑ "The Ile93Met mutation in the ubiquitin carboxy-terminal-hydrolase-L1 gene is not observed in European cases with familial Parkinson's disease". Neuroscience Letters 270 (1): 1–4. Jul 1999. doi:10.1016/s0304-3940(99)00465-6. PMID 10454131.
- ↑ "ACT and UCH-L1 polymorphisms in Parkinson's disease and age of onset". Movement Disorders 17 (4): 767–71. Jul 2002. doi:10.1002/mds.10179. PMID 12210873.
- ↑ "The S18Y polymorphic variant of UCH-L1 confers an antioxidant function to neuronal cells". Human Molecular Genetics 17 (14): 2160–71. Jul 2008. doi:10.1093/hmg/ddn115. PMID 18411255.
- ↑ 8.0 8.1 "Ubiquitin carboxy-terminal hydrolase L1 binds to and stabilizes monoubiquitin in neuron". Human Molecular Genetics 12 (16): 1945–58. Aug 2003. doi:10.1093/hmg/ddg211. PMID 12913066.
- ↑ "Ubiquitin hydrolase Uch-L1 rescues beta-amyloid-induced decreases in synaptic function and contextual memory". Cell 126 (4): 775–88. Aug 2006. doi:10.1016/j.cell.2006.06.046. PMID 16923396.
- ↑ "Ubiquitin C-terminal hydrolase L1 is required for pancreatic beta cell survival and function in lipotoxic conditions". Diabetologia 55 (1): 128–40. Jan 2012. doi:10.1007/s00125-011-2323-1. PMID 22038515.
- ↑ "Recessive loss of function of the neuronal ubiquitin hydrolase UCHL1 leads to early-onset progressive neurodegeneration". Proceedings of the National Academy of Sciences of the United States of America 110 (9): 3489–94. Feb 2013. doi:10.1073/pnas.1222732110. PMID 23359680. Bibcode: 2013PNAS..110.3489B.
- ↑ "Discovery of inhibitors that elucidate the role of UCH-L1 activity in the H1299 lung cancer cell line". Chemistry & Biology 10 (9): 837–46. Sep 2003. doi:10.1016/j.chembiol.2003.08.010. PMID 14522054.
- ↑ "The de-ubiquitinase UCH-L1 is an oncogene that drives the development of lymphoma in vivo by deregulating PHLPP1 and Akt signaling". Leukemia 24 (9): 1641–55. Sep 2010. doi:10.1038/leu.2010.138. PMID 20574456.
- ↑ "A new role for the neuronal ubiquitin C-terminal hydrolase-L1 (UCH-L1) in podocyte process formation and podocyte injury in human glomerulopathies". The Journal of Pathology 217 (3): 452–64. Feb 2009. doi:10.1002/path.2446. PMID 18985619.
- ↑ "UCH-L1 induces podocyte hypertrophy in membranous nephropathy by protein accumulation". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1842 (7): 945–58. Jul 2014. doi:10.1016/j.bbadis.2014.02.011. PMID 24583340.
- ↑ Peterson, Ivars (2006-10-14). "Knots in proteins". Science News. http://www.sciencenews.org/articles/20061014/mathtrek.asp.
- ↑ "Intricate knots in proteins: Function and evolution". PLOS Computational Biology 2 (9): e122. Sep 2006. doi:10.1371/journal.pcbi.0020122. PMID 16978047. Bibcode: 2006PLSCB...2..122V.
- ↑ 18.0 18.1 "The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility". Cell 111 (2): 209–18. Oct 2002. doi:10.1016/s0092-8674(02)01012-7. PMID 12408865. http://infoscience.epfl.ch/record/142130.
- ↑ "Interaction and colocalization of PGP9.5 with JAB1 and p27(Kip1)". Oncogene 21 (19): 3003–10. May 2002. doi:10.1038/sj.onc.1205390. PMID 12082530.
- ↑ "Parkin-mediated K63-polyubiquitination targets ubiquitin C-terminal hydrolase L1 for degradation by the autophagy-lysosome system". Cellular and Molecular Life Sciences 72 (9): 1811–24. May 2015. doi:10.1007/s00018-014-1781-2. PMID 25403879.
Further reading
- "Genetic causes of Parkinson's disease: UCHL-1". Cell and Tissue Research 318 (1): 189–94. Oct 2004. doi:10.1007/s00441-004-0917-3. PMID 15221445.
- "Microsequences of 145 proteins recorded in the two-dimensional gel protein database of normal human epidermal keratinocytes". Electrophoresis 13 (12): 960–9. Dec 1992. doi:10.1002/elps.11501301199. PMID 1286667.
- "The gene for human neurone specific ubiquitin C-terminal hydrolase (UCHL1, PGP9.5) maps to chromosome 4p14". Annals of Human Genetics 55 (Pt 4): 273–8. Oct 1991. doi:10.1111/j.1469-1809.1991.tb00853.x. PMID 1840236.
- "Neuronal protein gene product 9.5 (IEF SSP 6104) is expressed in cultured human MRC-5 fibroblasts of normal origin and is strongly down-regulated in their SV40 transformed counterparts". FEBS Letters 280 (2): 235–40. Mar 1991. doi:10.1016/0014-5793(91)80300-R. PMID 1849484.
- "The structure of the human gene encoding protein gene product 9.5 (PGP9.5), a neuron-specific ubiquitin C-terminal hydrolase". The Biochemical Journal 268 (2): 521–4. Jun 1990. doi:10.1042/bj2680521. PMID 2163617.
- "Molecular cloning of cDNA coding for human PGP 9.5 protein. A novel cytoplasmic marker for neurones and neuroendocrine cells". FEBS Letters 210 (2): 157–60. Jan 1987. doi:10.1016/0014-5793(87)81327-3. PMID 2947814.
- "Isolation of PGP 9.5, a new human neurone-specific protein detected by high-resolution two-dimensional electrophoresis". Journal of Neurochemistry 40 (6): 1542–7. Jun 1983. doi:10.1111/j.1471-4159.1983.tb08124.x. PMID 6343558.
- "Human TRE17 oncogene is generated from a family of homologous polymorphic sequences by single-base changes". DNA and Cell Biology 12 (2): 107–18. Mar 1993. doi:10.1089/dna.1993.12.107. PMID 8471161.
- "Substrate binding and catalysis by ubiquitin C-terminal hydrolases: identification of two active site residues". Biochemistry 35 (21): 6735–44. May 1996. doi:10.1021/bi960099f. PMID 8639624.
- "Localization and characterization of white blood cell populations within the human ovary throughout the menstrual cycle and menopause". Human Reproduction 11 (4): 790–7. Apr 1996. doi:10.1093/oxfordjournals.humrep.a019256. PMID 8671330.
- "The immunolocalization of PGP 9.5 in normal human kidney and renal cell carcinoma". Il Giornale di Chirurgia 18 (10): 521–4. Oct 1997. PMID 9435142.
- "Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases". Biochemistry 37 (10): 3358–68. Mar 1998. doi:10.1021/bi972274d. PMID 9521656.
- "The ubiquitin pathway in Parkinson's disease". Nature 395 (6701): 451–2. Oct 1998. doi:10.1038/26652. PMID 9774100. Bibcode: 1998Natur.395..451L.
- "Cleavage of the C-terminus of NEDD8 by UCH-L3". Biochemical and Biophysical Research Communications 251 (3): 688–92. Oct 1998. doi:10.1006/bbrc.1998.9532. PMID 9790970.
- "Intron-exon structure of ubiquitin c-terminal hydrolase-L1". DNA Research 5 (6): 397–400. Dec 1998. doi:10.1093/dnares/5.6.397. PMID 10048490.
- "Low frequency of pathogenic mutations in the ubiquitin carboxy-terminal hydrolase gene in familial Parkinson's disease". NeuroReport 10 (2): 427–9. Feb 1999. doi:10.1097/00001756-199902050-00040. PMID 10203348.
- "The Ile93Met mutation in the ubiquitin carboxy-terminal-hydrolase-L1 gene is not observed in European cases with familial Parkinson's disease". Neuroscience Letters 270 (1): 1–4. Jul 1999. doi:10.1016/S0304-3940(99)00465-6. PMID 10454131.
- "Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice". Nature Genetics 23 (1): 47–51. Sep 1999. doi:10.1038/12647. PMID 10471497.
- "The ubiquitin carboxy-terminal hydrolase-L1 gene S18Y polymorphism does not confer protection against idiopathic Parkinson's disease". Neuroscience Letters 293 (2): 127–30. Oct 2000. doi:10.1016/S0304-3940(00)01510-X. PMID 11027850.
- "Alpha-synuclein has an altered conformation and shows a tight intermolecular interaction with ubiquitin in Lewy bodies". Acta Neuropathologica 102 (4): 329–34. Oct 2001. doi:10.1007/s004010100369. PMID 11603807.
External links
- Ubiquitin+Carboxy-Terminal+Hydrolase at the US National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P09936 (Ubiquitin carboxyl-terminal hydrolase isozyme L1) at the PDBe-KB.
 |


