Biology:Micrococcal nuclease
| Micrococcal nuclease | |||||||||
|---|---|---|---|---|---|---|---|---|---|
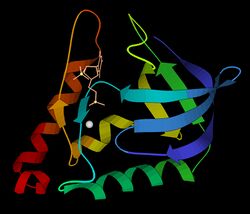 Ribbon schematic of micrococcal nuclease 3D structure, with Ca2+ and TdtP inhibitor | |||||||||
| Identifiers | |||||||||
| EC number | 3.1.31.1 | ||||||||
| CAS number | 9013-53-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
| Thermonuclease | |||||||
|---|---|---|---|---|---|---|---|
| Identifiers | |||||||
| Organism | |||||||
| Symbol | nuc | ||||||
| UniProt | P00644 | ||||||
| |||||||
| Staphylococcal nuclease | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| Symbol | ? | ||||||||
| Pfam | PF00565 | ||||||||
| Pfam clan | CL0049 | ||||||||
| InterPro | IPR016071 | ||||||||
| PROSITE | PDOC00865 | ||||||||
| CATH | 1tt2 | ||||||||
| SCOP2 | 1tt2 / SCOPe / SUPFAM | ||||||||
| |||||||||
Micrococcal nuclease (EC 3.1.31.1, S7 Nuclease, MNase, spleen endonuclease, thermonuclease, nuclease T, micrococcal endonuclease, nuclease T', staphylococcal nuclease, spleen phosphodiesterase, Staphylococcus aureus nuclease, Staphylococcus aureus nuclease B, ribonucleate (deoxynucleate) 3'-nucleotidohydrolase) is an endo-exonuclease that preferentially digests single-stranded nucleic acids. The rate of cleavage is 30 times greater at the 5' side of A or T than at G or C and results in the production of mononucleotides and oligonucleotides with terminal 3'-phosphates. The enzyme is also active against double-stranded DNA and RNA and all sequences will be ultimately cleaved.
Characteristics
The enzyme has a molecular weight of 16.9kDa.
The pH optimum is reported as 9.2. The enzyme activity is strictly dependent on Ca2+ and the pH optimum varies according to Ca2+ concentration.[1] The enzyme is therefore easily inactivated by EGTA.
Sources
This enzyme is the extracellular nuclease of Staphylococcus aureus. Two strains, V8 and Foggi, yield almost identical enzymes.[2] A common source is E.coli cells carrying a cloned nuc gene encoding Staphylococcus aureus extracellular nuclease (micrococcal nuclease).
Structure
The 3-dimensional structure of micrococcal nuclease (then called Staphyloccal nuclease) was solved very early in the history of protein crystallography, in 1969,[3] deposited as now-obsolete Protein Data Bank file 1SNS. Higher-resolution, more recent crystal structures are available for the apo form as Protein Data Bank file 1SNO: [1] and for the thymidine-diphosphate-inhibited form as Protein Data Bank file 3H6M: [2] or 1SNC: [3]. As seen in the ribbon diagram above, the nuclease molecule has 3 long alpha helices and a 5-stranded, barrel-shaped beta sheet, in an arrangement known as the OB-fold (for oligonucleotide-binding fold) as classified in the SCOP database.
Applications
- CUT&RUN sequencing, antibody-targeted controlled cleavage by micrococcal nuclease for transcriptomic profiling.
- Hydrolysis of nucleic acids in crude cell-free extracts.
- Sequencing of RNA.
- Preparation of rabbit reticulocyte lysates.
- Studies of chromatin structure.
- Removal of nucleic acids from laboratory protein preparations allowing for protein folding and structure-function studies.
- Research on the mechanisms of protein folding.
See also
References
- ↑ "Characterization of a nuclease produced by Staphylococcus aureus". J. Biol. Chem. 242 (5): 1016–20. 1967. doi:10.1016/S0021-9258(18)96225-3. PMID 6020427.
- ↑ "Staphylococcal nuclease (Foggi strain). I. Order of cyanogen bromide fragments and a "fourth" histidine residue". J. Biol. Chem. 243 (18): 4769–77. 1968. doi:10.1016/S0021-9258(18)93185-6. PMID 5687719.
- ↑ "A High Resolution Structure of an Inhibitor Complex of the Extracellular Nuclease of Staphylococcus aureus: I. Experimental Procedures and Chain Tracing". J. Biol. Chem. 246 (7): 2303–2316. 1971. doi:10.1016/S0021-9258(19)77221-4. PMID 5555571.
- http://www.thermoscientificbio.com/dna-and-rna-modifying-enzymes/micrococcal-nuclease/
- http://www.worthington-biochem.com/NFCP/default.html
- http://www.thermoscientificbio.com/uploadedFiles/Resources/en0181-usa-msds.pdf - A material and safety data sheet for the product
- http://www.thermoscientificbio.com/uploadedFiles/Resources/en018-product-information.pdf - A Product Information sheet
External links
- Micrococcal+Nuclease at the US National Library of Medicine Medical Subject Headings (MeSH)
- EC 3.1.31.1
 |
