Biology:RNA-induced silencing complex
The RNA-induced silencing complex, or RISC, is a multiprotein complex, specifically a ribonucleoprotein, which functions in gene silencing via a variety of pathways at the transcriptional and translational levels.[1] Using single-stranded RNA (ssRNA) fragments, such as microRNA (miRNA), or double-stranded small interfering RNA (siRNA), the complex functions as a key tool in gene regulation.[2] The single strand of RNA acts as a template for RISC to recognize complementary messenger RNA (mRNA) transcript. Once found, one of the proteins in RISC, Argonaute, activates and cleaves the mRNA. This process is called RNA interference (RNAi) and it is found in many eukaryotes; it is a key process in defense against viral infections, as it is triggered by the presence of double-stranded RNA (dsRNA).[3][4][1]
Discovery
The biochemical identification of RISC was conducted by Gregory Hannon and his colleagues at the Cold Spring Harbor Laboratory.[5] This was only a couple of years after the discovery of RNA interference in 1998 by Andrew Fire and Craig Mello, who shared the 2006 Nobel Prize in Physiology or Medicine.[3]

Hannon and his colleagues attempted to identify the RNAi mechanisms involved in gene silencing, by dsRNAs, in Drosophila cells. Drosophila S2 cells were transfected with a lacZ expression vector to quantify gene expression with β-galactosidase activity. Their results showed co-transfection with lacZ dsRNA significantly reduced β-galactosidase activity compared to control dsRNA. Therefore, dsRNAs control gene expression via sequence complementarity.
S2 cells were then transfected with Drosophila cyclin E dsRNA. Cycline E is an essential gene for cell cycle progression into the S phase. Cyclin E dsRNA arrested the cell cycle at the G1 phase (before the S phase). Therefore, RNAi can target endogenous genes.
In addition, cyclin E dsRNA only diminished cyclin E RNA — a similar result was also shown using dsRNA corresponding to cyclin A which acts in S, G2 and M phases of the cell cycle. This shows the characteristic hallmark of RNAi: the reduced levels of mRNAs correspond to the levels of dsRNA added.
To test whether their observation of decreased mRNA levels was a result of mRNA being targeted directly (as suggested by data from other systems), Drosophila S2 cells were transfected with either Drosophila cyclin E dsRNAs or lacZ dsRNAs and then incubated with synthetic mRNAs for cyclin E or lacZ.
Cells transfected with cyclin E dsRNAs only showed degradation in cyclin E transcripts — the lacZ transcripts were stable. Conversely, cells transfected with lacZ dsRNAs only showed degradation in lacZ transcripts and not cyclin E transcripts. Their results led Hannon and his colleagues to suggest RNAi degrades target mRNA through a 'sequence-specific nuclease activity'. They termed the nuclease enzyme RISC.[5] Later Devanand Sarkar and his colleagues Prasanna K. Santhekadur and Byoung Kwon Yoo at the Virginia Commonwealth University elucidated the RISC activity and its molecular mechanism in cancer cells and they identified another new component of the RISC, called AEG-1 [47].
Function in RNA interference
Incorporation of siRNA/miRNA
The RNase III Dicer is a critical member of RISC that initiates the RNA interference process by producing double-stranded siRNA or single-stranded miRNA. Enzymatic cleavage of dsRNA within the cell produces the short siRNA fragments of 21-23 nucleotides in length with a two-nucleotide 3' overhang.[6][7] Dicer also processes pre-miRNA, which forms a hairpin loop structure to mimic dsRNA, in a similar fashion. dsRNA fragments are loaded into RISC with each strand having a different fate based on the asymmetry rule phenomenon, the selection of one strand as the guide strand over the other based on thermodynamic stability.[8][9][10][11] The newly generated miRNA or siRNA act as single-stranded guide sequences for RISC to target mRNA for degradation.[12][13]
- The strand with the less thermodynamically stable 5' end is selected by the protein Argonaute and integrated into RISC.[11][14] This strand is known as the guide strand and targets mRNA for degradation.
- The other strand, known as the passenger strand, is degraded by RISC.[15]

Gene regulation

Major proteins of RISC, Ago2, SND1, and AEG-1, act as crucial contributors to the gene silencing function of the complex.[16]
RISC uses the guide strand of miRNA or siRNA to target complementary 3'-untranslated regions (3'UTR) of mRNA transcripts via Watson-Crick base pairing, allowing it to regulate gene expression of the mRNA transcript in a number of ways.[17][1]
mRNA degradation
The most understood function of RISC is degradation of target mRNA which reduces the levels of transcript available to be translated by ribosomes. The endonucleolytic cleavage of the mRNA complementary to the RISC's guide strand by Argonaute protein is the key to RNAi initiation.[18] There are two main requirements for mRNA degradation to take place:
- a near-perfect complementary match between the guide strand and target mRNA sequence, and,
- a catalytically active Argonaute protein, called a 'slicer', to cleave the target mRNA.[1]
There are two major pathways of mRNA degradation once cleavage has occurred. Both are initiated through degradation of the mRNA's poly(A) tail, resulting in removal of the mRNA's 5' cap.
- 5'-to-3' degradation of the transcript occurs by XRN1 exonuclease in cytoplasmic bodies called P-bodies.[19]
- 3'-to-5' degradation of the transcript is conducted by the exosome and Ski complex.[18]
Translational repression
RISC can modulate the loading of ribosome and accessory factors in translation to repress expression of the bound mRNA transcript. Translational repression only requires a partial sequence match between the guide strand and target mRNA.[1]
Translation can be regulated at the initiation step by:
- preventing the binding of the eukaryotic translation initiation factor (eIF) to the 5' cap. It has been noted RISC can deadenylate the 3' poly(A) tail which might contribute to repression via the 5' cap.[2][17]
- preventing the binding of the 60S ribosomal subunit binding to the mRNA can repress translation.[20]
Translation can be regulated at post-initiation steps by:
- peptide degradation,
- promoting premature termination of translation ribosomes,[21] or,
- slowing elongation.[22]
There is still speculation on whether translational repression via initiation and post-initiation is mutually exclusive.
Heterochromatin formation
Some RISCs are able to directly target the genome by recruiting histone methyltransferases to form heterochromatin at the gene locus, silencing the gene. These RISCs take the form of a RNA-induced transcriptional silencing complex (RITS). The best studied example is with the yeast RITS.[1][23][24]
RITS has been shown to direct heterochromatin formation at centromeres through recognition of centromeric repeats. Through base-pairing of siRNA (guide strand) to target chromatin sequences, histone-modifying enzymes can be recruited.[25]
The mechanism is not well understood; however, RITS degrade nascent mRNA transcripts. It has been suggested this mechanism acts as a 'self-reinforcing feedback loop' as the degraded nascent transcripts are used by RNA-dependent RNA polymerase (RdRp) to generate more siRNAs.[26]
In Schizosaccharomyces pombe and Arabidopsis, the processing of dsRNA targets into siRNA by Dicer RNases can initiate a gene silencing pathway by heterochromatin formation. An Argonaute protein known as AGO4 interacts with the small RNAs that define heterochromatic sequences. A histone methyl transferase (HMT), H3K9, methylates histone H3 and recruits chromodomain proteins to the methylation sites. DNA methylation maintains the silencing of genes as the heterochromatin sequences can be established or spread.[27]
DNA elimination
The siRNA generated by RISCs seem to have a role in degrading DNA during somatic macronucleus development in protozoa Tetrahymena. It is similar to the epigenetic control of heterochromatin formation and is implied as a defense against invading genetic elements.[27]
Similar to heterochromatin formation in S. pombe and Arabidopsis, a Tetrahymena protein related to the Argonaute family, Twi1p, catalyzes DNA elimination of target sequences known as internal elimination sequences (IESs). Using methyltransferases and chromodomain proteins, IESs are heterochromatized and eliminated from the DNA.[27]
RISC-associated proteins
The complete structure of RISC is still unsolved. Many studies have reported a range of sizes and components for RISC but it is not entirely sure whether this is due to there being a number of RISC complexes or due to the different sources that different studies use.[28]
| Complex | Source | Known/apparent components | Estimated size | Apparent function in RNAi pathway |
|---|---|---|---|---|
| Dcr2-R2D2[29] | D. melanogaster S2 cells | Dcr2, R2D2 | ~250 kDa | dsRNA processing, siRNA binding |
| RLC (A)[30][31] | D. melanogaster embryos | Dcr2, R2D2 | NR | dsRNA processing, siRNA binding, precursor to RISC |
| Holo-RISC[30][31] | D. melanogaster embryos | Ago 2, Dcr1, Dcr2, Fmr1/Fxr, R2D2, Tsn, Vig | ~80S | Target-RNA binding and cleavage |
| RISC[32] | D. melanogaster S2 cells | Ago2 | ~140 kDa | Target-RNA binding and cleavage |
| Fmr1-associated complex[33] | D. melanogaster S2 cells | L5, L11, 5S rRNA, Fmr1/Fxr, Ago2, Dmp68 | NR | Possible target-RNA binding and cleavage |
| Minimal RISC[34][35][36][37] | HeLa cells | eIF2C1 (Ago1) or eIF2C2 (Ago2) | ~160 kDa | Target-RNA binding and cleavage |
| miRNP[38][39] | HeLa cells | eIF2C2 (ago2), Gemin3, Gemin4 | ~550 kDa | miRNA association, target-RNA binding and cleavage |
Ago, Argonaute; Dcr, Dicer; Dmp68, D. melanogaster orthologue of mammalian p68 RNA unwindase; eIF2C1, eukaryotic translation initiation factor 2C1; eIF2C2, eukaryotic translation initiation factor 2C2; Fmr1/Fxr, D. melanogaster orthologue of the fragile-X mental retardation protein; miRNP, miRNA-protein complex; NR, not reported; Tsn, Tudor-staphylococcal nuclease; Vig, vasa intronic gene.
Regardless, it is apparent that Argonaute proteins are present and are essential for function. Furthermore, there are insights into some of the key proteins (in addition to Argonaute) within the complex, which allow RISC to carry out its function.
Argonaute proteins
Argonaute proteins are a family of proteins found in prokaryotes and eukaryotes. Their function in prokaryotes is unknown but in eukaryotes they are responsible for RNAi.[40] There are eight family members in human Argonautes of which only Argonaute 2 is exclusively involved in targeted RNA cleavage in RISC.[37]
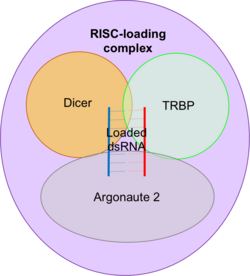
RISC-loading complex
The RISC-loading complex (RLC) is the essential structure required to load dsRNA fragments into RISC in order to target mRNA. The RLC consists of dicer, the transactivating response RNA-binding protein (TRBP) and Argonaute 2.
- Dicer is an RNase III endonuclease which generates the dsRNA fragments to be loaded that direct RNAi.
- TRBP is a protein with three double-stranded RNA-binding domains.
- Argonaute 2 is an RNase and is the catalytic centre of RISC.
Dicer associates with TRBP and Argonaute 2 to facilitate the transfer of the dsRNA fragments generated by Dicer to Argonaute 2.[41][42]
More recent research has shown the human RNA helicase A could help facilitate the RLC.[43]
Other proteins
Recently identified members of RISC are SND1 and MTDH.[44] SND1 and MTDH are oncogenes and regulate various gene expression.[45]
| Protein | Species the protein is found |
|---|---|
| Dcr1[30] | D. melanogaster |
| Dcr2[29][30][31] | D. melanogaster |
| R2D2[30][31] | D. melanogaster |
| Ago2[30][46][32][33] | D. melanogaster |
| Dmp68[33] | D. melanogaster |
| Fmr1/Fxr[30][47][33] | D. melanogaster |
| Tsn[30][48] | D. melanogaster |
| Vig[30][47] | D. melanogaster |
| Polyribosomes, ribosome components[5][30][46][33][49] | D. melanogaster, T. brucei |
| eIF2C1 (Ago1)[34] | H. sapiens |
| eIF2C2 (Ago2)[34][35][37][39] | H. sapiens |
| Gemin3[38][39] | H. sapiens |
| Gemin4[38][39] | H. sapiens |
Ago, Argonaute; Dcr, Dicer; Dmp68, D. melanogaster orthologue of mammalian p68 RNA unwindase; eIF2C1, eukaryotic translation initiation factor 2C1; eIF2C2, eukaryotic translation initiation factor 2C2; Fmr1/Fxr, D. melanogaster orthologue of the fragile-X mental retardation protein; Tsn, Tudor-staphylococcal nuclease; Vig, vasa intronic gene.
Binding of mRNA
It is as yet unclear how the activated RISC complex locates the mRNA targets in the cell, though it has been shown that the process can occur in situations outside of ongoing protein translation from mRNA.[50]
Endogenously expressed miRNA in metazoans is usually not perfectly complementary to a large number of genes and thus, they modulate expression via translational repression.[51][52] However, in plants, the process has a much greater specificity to target mRNA and usually each miRNA only binds to one mRNA. A greater specificity means mRNA degradation is more likely to occur.[53]
See also
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 "The RNA-induced silencing complex: A versatile gene-silencing machine". Journal of Biological Chemistry 284 (27): 17897–17901. 2009. doi:10.1074/jbc.R900012200. PMID 19342379.
- ↑ 2.0 2.1 "Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?". Nature Reviews Genetics 9 (2): 102–114. 2008. doi:10.1038/nrg2290. PMID 18197166. https://escholarship.mcgill.ca/concern/articles/cz30pz55f.
- ↑ 3.0 3.1 "Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans". Nature 391 (6669): 806–811. 1998. doi:10.1038/35888. PMID 9486653. Bibcode: 1998Natur.391..806F.
- ↑ Watson, James D. (2008). Molecular Biology of the Gene. San Francisco, CA: Cold Spring Harbor Laboratory Press. pp. 641–648. ISBN 978-0-8053-9592-1.
- ↑ 5.0 5.1 5.2 "An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells". Nature 404 (6775): 293–296. 2000. doi:10.1038/35005107. PMID 10749213. Bibcode: 2000Natur.404..293H.
- ↑ "RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals". Cell 101 (1): 25–33. 2000. doi:10.1016/S0092-8674(00)80620-0. PMID 10778853.
- ↑ "The contributions of dsRNA structure to Dicer specificity and efficiency". RNA 11 (5): 674–682. 2005. doi:10.1261/rna.7272305. PMID 15811921.
- ↑ Hutvagner, Gyorgy (2005). "Small RNA asymmetry in RNAi: Function in RISC assembly and gene regulation" (in en). FEBS Letters 579 (26): 5850–5857. doi:10.1016/j.febslet.2005.08.071. ISSN 1873-3468. PMID 16199039.
- ↑ "Asymmetry in the assembly of the RNAi enzyme complex". Cell 115 (2): 199–208. 2003. doi:10.1016/S0092-8674(03)00759-1. PMID 14567917.
- ↑ "Functional siRNAs and miRNAs exhibit strand bias". Cell 115 (2): 209–216. 2003. doi:10.1016/S0092-8674(03)00801-8. PMID 14567918.
- ↑ 11.0 11.1 "On the road to reading the RNA-interference code". Nature 457 (7228): 396–404. 2009. doi:10.1038/nature07754. PMID 19158785. Bibcode: 2009Natur.457..396S.
- ↑ Preall, Jonathan B.; Sontheimer, Erik J. (2005-11-18). "RNAi: RISC Gets Loaded" (in en). Cell 123 (4): 543–545. doi:10.1016/j.cell.2005.11.006. ISSN 0092-8674. PMID 16286001.
- ↑ "RNA interference overview | Abcam". https://www.abcam.com/pathways/rna-interference---a-comprehensive-overview.
- ↑ Preall, Jonathan B.; He, Zhengying; Gorra, Jeffrey M.; Sontheimer, Erik J. (2006-03-07). "Short Interfering RNA Strand Selection Is Independent of dsRNA Processing Polarity during RNAi in Drosophila" (in English). Current Biology 16 (5): 530–535. doi:10.1016/j.cub.2006.01.061. ISSN 0960-9822. PMID 16527750.
- ↑ "Human RISC couples microRNA biogenesis and posttranscriptional gene silencing". Cell 123 (4): 631–640. 2005. doi:10.1016/j.cell.2005.10.022. PMID 16271387.
- ↑ Santhekadur, Prasanna K.; Kumar, Divya P. (2020-06-01). "RISC assembly and post-transcriptional gene regulation in Hepatocellular Carcinoma" (in en). Genes & Diseases 7 (2): 199–204. doi:10.1016/j.gendis.2019.09.009. ISSN 2352-3042. PMID 32215289.
- ↑ 17.0 17.1 "Let-7 microRNA-mediated mRNA deadenylation and translational repression in a mammalian cell-free system". Genes & Development 21 (15): 1857–1862. 2007. doi:10.1101/gad.1566707. PMID 17671087.
- ↑ 18.0 18.1 ORBAN, TAMAS I.; IZAURRALDE, ELISA (April 2005). "Decay of mRNAs targeted by RISC requires XRN1, the Ski complex, and the exosome". RNA 11 (4): 459–469. doi:10.1261/rna.7231505. ISSN 1355-8382. PMID 15703439.
- ↑ "Argonaute2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies". Nature Cell Biology 7 (6): 633–636. 2005. doi:10.1038/ncb1265. PMID 15908945.
- ↑ "MicroRNA silencing through RISC recruitment of eIF6". Nature 447 (7146): 823–828. 2007. doi:10.1038/nature05841. PMID 17507929. Bibcode: 2007Natur.447..823C.
- ↑ "Short RNAs repress translation after initiation in mammalian cells". Molecular Cell 21 (4): 533–542. 2006. doi:10.1016/j.molcel.2006.01.031. PMID 16483934.
- ↑ "Evidence that microRNAs are associated with translating messenger RNAs in human cells". Nature Structural & Molecular Biology 13 (12): 1102–1107. 2006. doi:10.1038/nsmb1174. PMID 17128271.
- ↑ "RNAi-mediated targeting of heterchromatin by the RITS complex". Science 303 (5658): 672–676. 2004. doi:10.1126/science.1093686. PMID 14704433. Bibcode: 2004Sci...303..672V.
- ↑ "RITS acts in cis to promote RNA interference-mediated transcription and post-transcriptional silencing". Nature Genetics 36 (11): 1174–1180. 2004. doi:10.1038/ng1452. PMID 15475954.
- ↑ Shimada, Yukiko; Mohn, Fabio; Bühler, Marc (2016-12-01). "The RNA-induced transcriptional silencing complex targets chromatin exclusively via interacting with nascent transcripts". Genes & Development 30 (23): 2571–2580. doi:10.1101/gad.292599.116. ISSN 0890-9369. PMID 27941123.
- ↑ "RNA-dependent RNA polymerase is an essential component of a self-enforcing loop coupling heterochromatin assembly to siRNA production". Proceedings of the National Academy of Sciences of the United States of America 102 (1): 152–157. 2005. doi:10.1073/pnas.0407641102. PMID 15615848.
- ↑ 27.0 27.1 27.2 "Small RNAs in genome arrangement in Tetrahymena". Current Opinion in Genetics & Development 14 (2): 181–187. 2004. doi:10.1016/j.gde.2004.01.004. PMID 15196465.
- ↑ 28.0 28.1 28.2 Sontheimer EJ (2005). "Assembly and function of RNA silencing complexes". Nature Reviews Molecular Cell Biology 6 (2): 127–138. doi:10.1038/nrm1568. PMID 15654322.
- ↑ 29.0 29.1 "R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway". Science 301 (5641): 1921–1925. 2003. doi:10.1126/science.1088710. PMID 14512631. Bibcode: 2003Sci...301.1921L.
- ↑ 30.00 30.01 30.02 30.03 30.04 30.05 30.06 30.07 30.08 30.09 "A Dicer-2-dependent 80S complex cleaves targeted mRNAs during RNAi in Drosophila". Cell 117 (1): 83–94. 2004. doi:10.1016/S0092-8674(04)00258-2. PMID 15066284.
- ↑ 31.0 31.1 31.2 31.3 "RISC assembly defects in the Drosophila RNAi mutant armitage". Cell 116 (6): 831–841. 2004. doi:10.1016/S0092-8674(04)00218-1. PMID 15035985.
- ↑ 32.0 32.1 "Biochemical identification of Argonaute 2 as the sole protein required for RNA-induced silencing complex activity". Proceedings of the National Academy of Sciences of the United States of America 101 (40): 14385–14389. 2004. doi:10.1073/pnas.0405913101. PMID 15452342. Bibcode: 2004PNAS..10114385R.
- ↑ 33.0 33.1 33.2 33.3 33.4 "A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins". Genes & Development 16 (19): 2497–2508. 2002. doi:10.1101/gad.1022002. PMID 12368261.
- ↑ 34.0 34.1 34.2 "Single-stranded antisense siRNAs guide target RNA cleavage in RNAi". Cell 110 (5): 563–574. 2002. doi:10.1016/S0092-8674(02)00908-X. PMID 12230974.
- ↑ 35.0 35.1 "Argonaute2 is the catalytic engine of mammalian RNAi". Science 305 (5689): 1437–1441. 2004. doi:10.1126/science.1102513. PMID 15284456. Bibcode: 2004Sci...305.1437L.
- ↑ "RISC is a 5′ phosphomonoester-producing RNA endonuclease". Genes & Development 18 (9): 975–980. 2004. doi:10.1101/gad.1187904. PMID 15105377.
- ↑ 37.0 37.1 37.2 "Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs". Molecular Cell 15 (2): 1403–1408. 2004. doi:10.1016/j.molcel.2004.07.007. PMID 15260970.
- ↑ 38.0 38.1 38.2 "miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs". Genes & Development 16 (6): 720–728. 2002. doi:10.1101/gad.974702. PMID 11914277.
- ↑ 39.0 39.1 39.2 39.3 "A microRNA in a multiple-turnover RNAi enzyme complex". Science 297 (5589): 2056–2060. 2002. doi:10.1126/science.1073827. PMID 12154197. Bibcode: 2002Sci...297.2056H.
- ↑ Hall TM (2005). "Structure and function of Argonaute proteins". Cell 13 (10): 1403–1408. doi:10.1016/j.str.2005.08.005. PMID 16216572.
- ↑ "TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing". Nature 436 (7051): 740–744. 2005. doi:10.1038/nature03868. PMID 15973356. Bibcode: 2005Natur.436..740C.
- ↑ "Structural insights into RNA processing by the human RISC-loading complex". Nature Structural & Molecular Biology 16 (11): 1148–1153. 2009. doi:10.1038/nsmb.1673. PMID 19820710.
- ↑ "Structural insights into RISC assembly facilitated by dsRNA-binding domains of human RNA helices A (DHX9)". Nucleic Acids Research 41 (5): 3457–3470. 2013. doi:10.1093/nar/gkt042. PMID 23361462.
- ↑ "Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma". Hepatology 53 (5): 1538–1548. 2011. doi:10.1002/hep.24216. PMID 21520169.
- ↑ "Astrocyte elevated gene (AEG-1): a multifunctional regulator of normal and abnormal physiology". Pharmacology & Therapeutics 130 (1): 1–8. 2011. doi:10.1016/j.pharmthera.2011.01.008. PMID 21256156.
- ↑ 46.0 46.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedHammond2001 - ↑ 47.0 47.1 Cite error: Invalid
<ref>tag; no text was provided for refs namedCaudy2002 - ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedCaudy2003 - ↑ "An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei". RNA 9 (7): 802–808. 2003. doi:10.1261/rna.5270203. PMID 12810914.
- ↑ "mRNA translation is not a prerequisite for small interfering RNA-mediated mRNA cleavage". Differentiation 73 (6): 287–293. 2005. doi:10.1111/j.1432-0436.2005.00029.x. PMID 16138829.
- ↑ "Anti-viral RNA silencing: do we look like plants?". Retrovirology 3: 3. 2006. doi:10.1186/1742-4690-3-3. PMID 16409629.
- ↑ Bartel DP (2009). "MicroRNAs: target recognition and regulatory functions". Cell 136 (2): 215–233. doi:10.1016/j.cell.2009.01.002. PMID 19167326.
- ↑ "MicroRNAs and their regulator roles in plants". Annual Review of Plant Biology 57: 19–53. 2006. doi:10.1146/annurev.arplant.57.032905.105218. PMID 16669754.
Further reading
- Sontheimer, EJ (2005). "Assembly and function of RNA silencing complexes". Nature Reviews Molecular Cell Biology 6 (2): 127–138. doi:10.1038/nrm1568. PMID 15654322.
- "Structural insights into RISC assembly facilitated by dsRNA-binding domains of human RNA helicase A (DHX9)". Nucleic Acids Research 41 (5): 3457–70. March 2013. doi:10.1093/nar/gkt042. PMID 23361462.
- "The RNA-induced silencing complex is a Mg2+-dependent endonuclease". Current Biology 14 (9): 787–91. 2004. doi:10.1016/j.cub.2004.03.008. PMID 15120070.
External links
- RNA-Induced+Silencing+Complex at the US National Library of Medicine Medical Subject Headings (MeSH)
 |
