Medicine:Escherichia coli O157:H7
| Escherichia coli O157:H7 | |
|---|---|
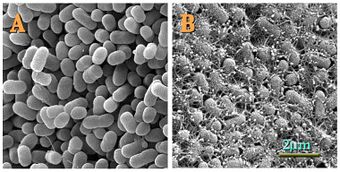 | |
| Topographical images of colonies of E. coli O157:H7 strains (A) 43895OW (curli non-producing) and (B) 43895OR (curli producing) grown on agar for 48 h at 28°C | |
| Specialty | Infectious disease |
Escherichia coli O157:H7 is a serotype of the bacterial species Escherichia coli and is one of the Shiga-like toxin–producing types of E. coli. It is a cause of disease, typically foodborne illness, through consumption of contaminated and raw food, including raw milk and undercooked ground beef.[1][2] Infection with this type of pathogenic bacteria may lead to hemorrhagic diarrhea, and to kidney failure; these have been reported to cause the deaths of children younger than five years of age, of elderly patients, and of patients whose immune systems are otherwise compromised.
Transmission is via the fecal–oral route, and most illness has been through distribution of contaminated raw leaf green vegetables, undercooked meat and raw milk.[3]
Signs and symptoms
E. coli O157:H7 infection often causes severe, acute hemorrhagic diarrhea (although nonhemorrhagic diarrhea is also possible) and abdominal cramps. Usually little or no fever is present, and the illness resolves in 5 to 10 days.[4] It can also sometimes be asymptomatic.[5]
In some people, particularly children under five years of age, persons whose immunologies are otherwise compromised, and the elderly, the infection can cause hemolytic–uremic syndrome (HUS), in which the red blood cells are destroyed and the kidneys fail. About 2–7% of infections lead to this complication. In the United States, HUS is the principal cause of acute kidney failure in children, and most cases of HUS are caused by E. coli O157:H7.[citation needed]
Bacteriology

Like the other strains of the E. coli, O157:H7 is gram-negative and oxidase-negative. Unlike many other strains, it does not ferment sorbitol, which provides a basis for clinical laboratory differentiation of the strain. Strains of E. coli that express Shiga and Shiga-like toxins gained that ability via infection with a prophage containing the structural gene coding for the toxin, and nonproducing strains may become infected and produce shiga-like toxins after incubation with shiga toxin positive strains. The prophage responsible seems to have infected the strain's ancestors fairly recently, as viral particles have been observed to replicate in the host if it is stressed in some way (e.g. antibiotics).[6][7]
All clinical isolates of E. coli O157:H7 possess the plasmid pO157.[8] The periplasmic catalase is encoded on pO157 and may enhance the virulence of the bacterium by providing additional oxidative protection when infecting the host.[9] E. coli O157:H7 non-hemorrhagic strains are converted to hemorrhagic strains by lysogenic conversion after bacteriophage infection of non-hemorrhagic cells.[citation needed]
Natural habitat
While it is relatively uncommon, the E. coli serotype O157:H7 can naturally be found in the intestinal contents of some cattle, goats, and even sheep.[citation needed] The digestive tract of cattle lack the Shiga toxin receptor globotriaosylceramide, and thus, these can be asymptomatic carriers of the bacterium.[10] The prevalence of E. coli O157:H7 in North American feedlot cattle herds ranges from 0 to 60%.[11] Some cattle may also be so-called "super-shedders" of the bacterium. Super-shedders may be defined as cattle exhibiting rectoanal junction colonization and excreting >103 to 4 CFU g−1 feces. Super-shedders have been found to constitute a small proportion of the cattle in a feedlot (<10%) but they may account for >90% of all E. coli O157:H7 excreted.[12]
Transmission
Infection with E. coli O157:H7 can come from ingestion of contaminated food or water, or oral contact with contaminated surfaces. Examples of this can be undercooked ground beef but also leafy vegetables and raw milk. Fields often get contaminated with the bacterium through irrigation processes or contaminated water naturally entering the soil.[13] It is highly virulent, with a low infectious dose: an inoculation of fewer than 10 to 100 colony-forming units (CFU) of E. coli O157:H7 is sufficient to cause infection, compared to over a million CFU for other pathogenic E. coli strains.[14]
Diagnosis
A stool culture can detect the bacterium. The sample is cultured on sorbitol-MacConkey (SMAC) agar, or the variant cefixime potassium tellurite sorbitol-MacConkey agar (CT-SMAC[15]). On SMAC agar, O157:H7 colonies appear clear due to their inability to ferment sorbitol, while the colonies of the usual sorbitol-fermenting serotypes of E. coli appear red. Sorbitol nonfermenting colonies are tested for the somatic O157 antigen before being confirmed as E. coli O157:H7. Like all cultures, diagnosis is time-consuming with this method; swifter diagnosis is possible using quick E. coli DNA extraction method[16] plus polymerase chain reaction techniques. Newer technologies using fluorescent and antibody detection are also under development.[citation needed]
Prevention
Avoiding the consumption of, or contact with, unpasteurised dairy products, undercooked beef, uncleaned vegetables, and undisinfected water reduces the risk of an E. coli infection. Proper hand washing with water that has been treated with adequate levels of chlorine or other effective disinfectants after using the lavatory or changing a diaper, especially among children or those with diarrhea, reduces the risk of transmission.[17][18]
Surveillance
E. coli O157:H7 infection is a nationally reportable disease in the US, Great Britain, and Germany. It is also reportable in most states of Australia including Queensland.[citation needed][19]
Treatment
While fluid replacement and blood pressure support may be necessary to prevent death from dehydration, most patients recover without treatment in 5–10 days. There is no evidence that antibiotics improve the course of disease, and treatment with antibiotics may precipitate hemolytic–uremic syndrome (HUS).[20] The antibiotics are thought to trigger prophage induction, and the prophages released by the dying bacteria infect other susceptible bacteria, converting them into toxin-producing forms. Antidiarrheal agents, such as loperamide (imodium), should also be avoided as they may prolong the duration of the infection.[citation needed]
Certain novel treatment strategies, such as the use of anti-induction strategies to prevent toxin production[21] and the use of anti-Shiga toxin antibodies,[22] have also been proposed.
History
United States
The United States Department of Agriculture banned the sale of ground beef contaminated with the O157:H7 strain in 1994.[23]
Culture and society
Costs
The pathogen results in an estimated 2,100 hospitalizations annually in the United States. The illness is often misdiagnosed; therefore, expensive and invasive diagnostic procedures may be performed. Patients who develop HUS often require prolonged hospitalization, dialysis, and long-term followup.[24]
See also
- 1993 Jack in the Box E. coli outbreak
- 1996 Odwalla E. coli outbreak
- 2011 Germany E. coli O104:H4 outbreak
- Escherichia coli O104:H4
- Escherichia coli O121
- Food-induced purpura
- List of foodborne illness outbreaks
- Walkerton E. coli outbreak
References
- ↑ "Microbe Profile: Escherichia coli O157:H7 - notorious relative of the microbiologist's workhorse". Microbiology 163 (1): 1–3. January 2017. doi:10.1099/mic.0.000387. PMID 28218576. https://www.research.ed.ac.uk/portal/files/31527884/1_micro000387.pdf.
- ↑ "Enterohaemorrhagic Escherichia coli in human medicine". International Journal of Medical Microbiology 295 (6–7): 405–18. October 2005. doi:10.1016/j.ijmm.2005.06.009. PMID 16238016.
- ↑ "Reports of Selected E. coli Outbreak Investigations". CDC.gov. 2019-11-22. https://www.cdc.gov/ecoli/outbreaks.html.
- ↑ "Management strategies in the treatment of neonatal and pediatric gastroenteritis". Infection and Drug Resistance 6: 133–61. October 2013. doi:10.2147/IDR.S12718. PMID 24194646.
- ↑ "The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine". Infection and Immunity 74 (1): 615–24. January 2006. doi:10.1128/IAI.74.1.615-624.2006. PMID 16369018.
- ↑ "Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea". Science 226 (4675): 694–96. November 1984. doi:10.1126/science.6387911. PMID 6387911. Bibcode: 1984Sci...226..694O.
- ↑ "Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities". Infection and Immunity 53 (1): 135–40. July 1986. doi:10.1128/IAI.53.1.135-140.1986. PMID 3522426.
- ↑ "A brief overview of Escherichia coli O157:H7 and its plasmid O157". Journal of Microbiology and Biotechnology 20 (1): 5–14. January 2010. doi:10.4014/jmb.0908.08007. PMID 20134227.
- ↑ "KatP, a novel catalase-peroxidase encoded by the large plasmid of enterohaemorrhagic Escherichia coli O157:H7". Microbiology 142 ( Pt 11) (11): 3305–15. November 1996. doi:10.1099/13500872-142-11-3305. PMID 8969527.
- ↑ "Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins". Proceedings of the National Academy of Sciences of the United States of America 97 (19): 10325–29. September 2000. doi:10.1073/pnas.190329997. PMID 10973498. Bibcode: 2000PNAS...9710325P.
- ↑ "Evaluation of animal genetic and physiological factors that affect the prevalence of Escherichia coli O157 in cattle". PLOS ONE 8 (2): e55728. 2013. doi:10.1371/journal.pone.0055728. PMID 23405204. Bibcode: 2013PLoSO...855728J.
- ↑ "Super-shedding and the link between human infection and livestock carriage of Escherichia coli O157". Nature Reviews. Microbiology 6 (12): 904–12. December 2008. doi:10.1038/nrmicro2029. PMID 19008890.
- ↑ Scutti, Susan. "Why deadly E. coli loves leafy greens". CNN. https://www.cnn.com/2018/05/02/health/e-coli-lettuce-explainer/index.html.
- ↑ J.D. Greig, E.C.D. Todd, C. Bartleson, and B. Michaels. March 25, 2010. "Infective Doses and Pathen Carriage ", pp. 19–20, USDA 2010 Food Safety Education Conference.
- ↑ "MACCONKEY SORBITOL AGAR (CT-SMAC)". http://www.solabia.com/solabia/produitsDiagnostic.nsf/0/27DFE62DDA44AF3AC12574C7003A6377/$file/TDS_BK147_v5.pdf.
- ↑ "Quick E. coli DNA extraction filter paper card". http://www.fortiusbio.com/Food_Pathogen_E-coli_DNA_Extraction_Strip_Card.html.
- ↑ "Viruses, Bacteria, and Parasites in the Digestive Tract - Health Encyclopedia - University of Rochester Medical Center". https://www.urmc.rochester.edu/encyclopedia/content.aspx?ContentTypeID=90&ContentID=P02019.
- ↑ "Preventing Foodborne Illness: Escherichia coli O157:H7". https://wonder.cdc.gov/wonder/prevguid/p0000417/p0000417.asp#head009000000000000.
- ↑ "Journal". https://academic.oup.com/cid/article-abstract/69/3/428/5146342?redirectedFrom=fulltext.
- ↑ "Effect of subinhibitory concentrations of antibiotics on extracellular Shiga-like toxin I". Infection 20 (1): 25–29. 1992. doi:10.1007/BF01704889. PMID 1563808.
- ↑ "Paradigms of pathogenesis: targeting the mobile genetic elements of disease". Frontiers in Cellular and Infection Microbiology 2: 161. December 2012. doi:10.3389/fcimb.2012.00161. PMID 23248780.
- ↑ "Antibody therapy in the management of shiga toxin-induced hemolytic uremic syndrome". Clinical Microbiology Reviews 17 (4): 926–41, table of contents. October 2004. doi:10.1128/CMR.17.4.926-941.2004. PMID 15489355.
- ↑ "Ban on E. Coli in Ground Beef Is to Extend to 6 More Strains". The New York Times. September 12, 2011. https://www.nytimes.com/2011/09/13/business/federal-officials-extend-e-coli-ban.html?scp=2&sq=Jack%20in%20the%20Box&st=cse.
- ↑ Berkenpas, E.; Millard, P.; Pereira da Cunha, M. (2005-12-13). "Detection of Escherichia coli O157:H7 with langasite pure shear horizontal surface acoustic wave sensors" (in en). Biosensors and Bioelectronics 21 (12): 2255–2262. doi:10.1016/j.bios.2005.11.005. PMID 16356708.
External links
| Classification |
|---|
- Haemolytic Uraemic Syndrome Help (HUSH) – a UK based charity
- E. coli: Protecting yourself and your family from a sometimes deadly bacterium
- Escherichia coli O157:H7 genomes and related information at PATRIC, a Bioinformatics Resource Center funded by NIAID
- For more information about reducing your risk of foodborne illness, visit the US Department of Agriculture's Food Safety and Inspection Service website or The Partnership for Food Safety Education | Fight BAC!
- briandeer.com, report from The Sunday Times on a UK outbreak, May 17, 1998
- CBS5 report on September 2006 outbreak
bg:Escherichia coli O157:H7
