Biology:Proteobacteria
| Proteobacteria | |
|---|---|
| |
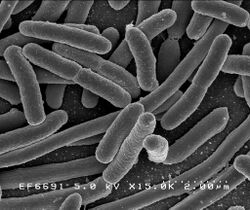
| Escherichia coli | |
| Scientific classification | |
| Domain: | Bacteria |
| Phylum: | Pseudomonadota Stackebrandt et al., 1988,[1] Garrity et al. 2005[2] |
| Classes | |
|
Alphaproteobacteria[3] | |
Proteobacteria is a major phylum of Gram-negative bacteria. They include a wide variety of pathogenic genera, such as Escherichia, Salmonella, Vibrio, Helicobacter, Yersinia, Legionellales, and many others.[10] Others are free-living (nonparasitic) and include many of the bacteria responsible for nitrogen fixation.
Carl Woese established this grouping in 1987, calling it informally the "purple bacteria and their relatives".[11] Because of the great diversity of forms found in this group, it was named after Proteus, a Greek god of the sea capable of assuming many different shapes and is not named after the Proteobacteria genus Proteus.[1][12]
Characteristics
All "Proteobacteria" are Gram-negative (though some may stain Gram-positive or Gram-variable in practice), with an outer membrane mainly composed of lipopolysaccharides. Many move about using flagella, but some are nonmotile or rely on bacterial gliding. The latter include the myxobacteria, an order of bacteria that can aggregate to form multicellular fruiting bodies. Also, a wide variety in the types of metabolism exists. Most members are facultatively or obligately anaerobic, chemolithoautotrophic, and heterotrophic, but numerous exceptions occur. A variety of genera, which are not closely related to each other, convert energy from light through photosynthesis and anoxygenic photosynthesis.
"Proteobacteria" are associated with the imbalance of microbiota of the lower reproductive tract of women. These species are associated with inflammation.[13]
Some Alphaproteobacteria can grow at very low levels of nutrients and have unusual morphology such as stalks and buds. Others include agriculturally important bacteria capable of inducing nitrogen fixation in symbiosis with plants. The type order is the Caulobacterales, comprising stalk-forming bacteria such as Caulobacter. The mitochondria of eukaryotes are thought to be descendants of an alphaproteobacterium.[14]
The Betaproteobacteria are highly metabolically diverse and contain chemolithoautotrophs, photoautotrophs, and generalist heterotrophs. The type order is the Burkholderiales, comprising an enormous range of metabolic diversity, including opportunistic pathogens.
The Gammaproteobacteria are the largest class in terms of species with validly published names. The type order is the Pseudomonadales, which include the genera Pseudomonas and the nitrogen-fixing Azotobacter.
The Deltaproteobacteria include bacteria that are predators on other bacteria and are important contributors to the anaerobic side of the sulfur cycle. The type order is the Myxococcales, which includes organisms with self-organising abilities such as Myxococcus spp.
The Epsilonproteobacteria are often slender, Gram-negative rods that are helical or curved. The type order is the Campylobacterales, which includes important food pathogens such as Campylobacter spp.
The Zetaproteobacteria are iron-oxidizing neutrophilic chemolithoautotrophs, distributed worldwide in estuaries and marine habitats. The type order is the Mariprofundales.
The Hydrogenophilalia are obligate thermophiles and include heterotrophs and autotrophs. The type order is the Hydrogenophilales.
The Acidithiobacillia contain only sulfur, iron, and uranium-oxidising autotrophs. The type order is the Acidithiobacillales, which includes economically important organisms used in the mining industry such as Acidithiobacillus spp.
The Oligoflexia are filamentous aerobes. The type order is the Oligoflexales, which contains the genus Oligoflexus.
Taxonomy
| Phylogeny of "Proteobacteria" | ||||||||||||||||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||
| Phylogeny of the "Proteobacteria" according to ARB living tree, iTOL, Bergey's and others. Acidobacteria (not to be confused with Acidithiobacillia) is used as outgroup. |
The group is defined primarily in terms of ribosomal RNA (rRNA) sequences. The "Proteobacteria" are divided into nine classes with validly published names, referred to by the Greek letters alpha through zeta, the Acidithiobacillia, Hydrogenophilalia, and Oligoflexia. These were previously regarded as subclasses of the phylum, but they are now treated as classes. These classes are monophyletic.[15][16][17] The genus Acidithiobacillus, part of the Gammaproteobacteria until it was transferred to class Acidithiobacillia in 2013,[18] was previously regarded as paraphyletic to the Betaproteobacteria according to multigenome alignment studies.[19] In 2017, the Betaproteobacteria was subject to major revisions and the class Hydrogenophilalia was created to contain the order Hydrogenophilales[20]
Proteobacterial classes with validly published names include some prominent genera:[21] e.g.:
- Alphaproteobacteria: Brucella, Rhizobium, Agrobacterium, Caulobacter, Rickettsia, Wolbachia, etc.
- Betaproteobacteria: Bordetella, Ralstonia, Neisseria, Nitrosomonas, etc.
- Gammaproteobacteria: Escherichia, Shigella, Salmonella, Yersinia, Buchnera, Haemophilus, Vibrio, Pseudomonas, etc.
- Deltaproteobacteria: Desulfovibrio, Geobacter, Bdellovibrio, etc.
- Epsilonproteobacteria: Helicobacter, Campylobacter, Wolinella, etc.
- Zetaproteobacteria: Mariprofundus, Ghiorsea
- Oligoflexia: Oligoflexus.
- Acidithiobacillia: Acidithiobacillus thiooxidans, Thermithiobacillus tepidarius
- Hydrogenophilalia: Hydrogenophilus thermoluteolus, Tepidiphilus margaritifer
Transformation
Transformation, a process in which genetic material passes from one bacterium to another,[22] has been reported in at least 30 species of "Proteobacteria" distributed in the classes alpha, beta, gamma and epsilon.[23] The best-studied "Proteobacteria" with respect to natural genetic transformation are the medically important human pathogens Neisseria gonorrhoeae (class beta), Haemophilus influenzae (class gamma) and Helicobacter pylori (class epsilon).[24] Natural genetic transformation is a sexual process involving DNA transfer from one bacterial cell to another through the intervening medium and the integration of the donor sequence into the recipient genome. In pathogenic "Proteobacteria", transformation appears to serve as a DNA repair process that protects the pathogen's DNA from attack by their host's phagocytic defenses that employ oxidative free radicals.[24]
References
- ↑ 1.0 1.1 Stackebrandt, E.; Murray, R. G. E.; Truper, H. G. (1988). "Proteobacteria classis nov., a Name for the Phylogenetic Taxon That Includes the "Purple Bacteria and Their Relatives"". International Journal of Systematic Bacteriology 38 (3): 321–325. doi:10.1099/00207713-38-3-321.
- ↑ Garrity, G. M., Bell, J. A. & Lilburn, T. (2005). Phylum XIV. Proteobacteria phyl. nov. In: Bergey’s Manual of Systematic Bacteriology, 2nd edn, vol. 2 (The Proteobacteria), part B (The Gammaproteobacteria), p. 1. Edited by D. J. Brenner, N. R. Krieg, J. T. Staley & G. M. Garrity. New York: Springer.
- ↑ "Class I. Alphaproteobacteria class. nov.". Bergey's Manual of Systematic Bacteriology Volume 2: The Proteobacteria Part C (The Alpha-, Beta-, Delta- and Epsilonproteobacteria (2nd ed.). Springer. 2005. p. 1. doi:10.1002/9781118960608.cbm00041. ISBN 9781118960608.
- ↑ 4.0 4.1 "Reclassification of Thiobacillus aquaesulis (Wood & Kelly, 1995) as Annwoodia aquaesulis gen. nov., comb. nov., transfer of Thiobacillus (Beijerinck, 1904) from the Hydrogenophilales to the Nitrosomonadales, proposal of Hydrogenophilalia class. nov. within the "Proteobacteria", and four new families within the orders Nitrosomonadales and Rhodocyclales.". International Journal of Systematic and Evolutionary Microbiology 67 (5): 1191–1205. 2017. doi:10.1099/ijsem.0.001927. PMID 28581923.
- ↑ 5.0 5.1 Williams KP, Kelly DP (2013). "Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria.". International Journal of Systematic and Evolutionary Microbiology 63 (Pt 8): 2901–2906. doi:10.1099/ijs.0.049270-0. PMID 23334881. https://semanticscholar.org/paper/fa17a78b8e9f295665d111a43414bc344ef68485.
- ↑ "Class IV. Deltaproteobacteria class. nov.". Bergey's Manual of Systematic Bacteriology Volume 2: The Proteobacteria Part C (The Alpha-, Beta-, Delta- and Epsilonproteobacteria (2nd ed.). Springer. 2005. p. 922. doi:10.1002/9781118960608.cbm00043. ISBN 9781118960608.
- ↑ "Class V. Epsilonproteobacteria class. nov.". Bergey's Manual of Systematic Bacteriology Volume 2: The Proteobacteria Part C (The Alpha-, Beta-, Delta- and Epsilonproteobacteria (2nd ed.). Springer. 2005. p. 1145. doi:10.1002/9781118960608.cbm00044. ISBN 9781118960608.
- ↑ Emerson, D.; Rentz, J. A.; Lilburn, T. G.; Davis, R. E.; Aldrich, H.; Chan, C.; Moyer, C. L. (2007). Reysenbach, Anna-Louise. ed. "A Novel Lineage of Proteobacteria Involved in Formation of Marine Fe-Oxidizing Microbial Mat Communities". PLOS ONE 2 (8): e667. doi:10.1371/journal.pone.0000667. PMID 17668050. Bibcode: 2007PLoSO...2..667E.
- ↑ Nakai R, Nishijima M, Tazato N, Handa Y, Karray F, Sayadi S, Isoda H, Naganuma T (2014). "Oligoflexus tunisiensis gen. nov., sp. nov., a Gram-negative, aerobic, filamentous bacterium of a novel proteobacterial lineage, and description of Oligoflexaceae fam. nov., Oligoflexales ord. nov. and Oligoflexia classis nov.". International Journal of Systematic and Evolutionary Microbiology 64 (Pt 10): 3353–3359. doi:10.1099/ijs.0.060798-0. PMID 25013226.
- ↑ Madigan, M. and J. Martinko. (eds.) (2005). Brock Biology of Microorganisms (11th ed.). Prentice Hall. ISBN 978-0-13-144329-7.
- ↑ Woese, CR (1987). "Bacterial evolution". Microbiological Reviews 51 (2): 221–71. doi:10.1128/MMBR.51.2.221-271.1987. PMID 2439888.
- ↑ "Proteobacteria". Discover Life: Tree of Life. http://stri.discoverlife.org/mp/20m?tree=Proteobacteria&res=800.
- ↑ Bennett, John (2015). Mandell, Douglas, and Bennett's principles and practice of infectious diseases. Philadelphia, PA: Elsevier/Saunders. ISBN 9781455748013; Access provided by the University of Pittsburgh
- ↑ Roger AJ, Muñoz-Gómez SA, Kamikawa R (2017). "The origin and diversification of mitochondria". Current Biology 27 (21): R1177–R1192. doi:10.1016/j.cub.2017.09.015. PMID 29112874.
- ↑ Noel R. Krieg; Don J. Brenner; James T. Staley (2005). Bergey's Manual of Systematic Bacteriology: The Proteobacteria. Springer. ISBN 978-0-387-95040-2.
- ↑ Ciccarelli, FD; Doerks, T; Von Mering, C; Creevey, CJ; Snel, B; Bork, P (2006). "Toward automatic reconstruction of a highly resolved tree of life". Science 311 (5765): 1283–7. doi:10.1126/science.1123061. PMID 16513982. Bibcode: 2006Sci...311.1283C.
- ↑ Yarza, P; Ludwig, W; Euzéby, J; Amann, R; Schleifer, KH; Glöckner, FO; Rosselló-Móra, R (2010). "Update of the All-Species Living Tree Project based on 16S and 23S rRNA sequence analyses". Systematic and Applied Microbiology 33 (6): 291–9. doi:10.1016/j.syapm.2010.08.001. PMID 20817437..
- ↑ Williams, KP; Kelly, DP (2013). "Proposal for a new class within the phylum Proteobacteria, Acidithiobacillia classis nov., with the type order Acidithiobacillales, and emended description of the class Gammaproteobacteria". International Journal of Systematic and Evolutionary Microbiology 63 (Pt 8): 2901–6. doi:10.1099/ijs.0.049270-0. PMID 23334881. https://semanticscholar.org/paper/fa17a78b8e9f295665d111a43414bc344ef68485.
- ↑ Williams, K. P.; Gillespie, J. J.; Sobral, B. W. S.; Nordberg, E. K.; Snyder, E. E.; Shallom, J. M.; Dickerman, A. W. (2010). "Phylogeny of Gammaproteobacteria". Journal of Bacteriology 192 (9): 2305–14. doi:10.1128/JB.01480-09. PMID 20207755.
- ↑ Boden, R.; Hutt, L. P.; Rae, A. W. (2017). "Reclassification of Thiobacillus aquaesulis (Wood & Kelly, 1995) as Annwoodia aquaesulis gen. nov., comb. nov., transfer of Thiobacillus (Beijerinck, 1904) from the Hydrogenophilales to the Nitrosomonadales, proposal of Hydrogenophilalia class. nov. within the Proteobacteria, and four new families within the orders Nitrosomonadales and Rhodocyclales.". International Journal of Systematic and Evolutionary Microbiology 67 (5): 1191–1205. doi:10.1099/ijsem.0.001927. PMID 28581923.
- ↑ Interactive Tree of Life
- ↑ "Bacterial transformation: distribution, shared mechanisms and divergent control". Nat. Rev. Microbiol. 12 (3): 181–96. 2014. doi:10.1038/nrmicro3199. PMID 24509783.
- ↑ "Natural genetic transformation: prevalence, mechanisms and function". Res. Microbiol. 158 (10): 767–78. 2007. doi:10.1016/j.resmic.2007.09.004. PMID 17997281.
- ↑ 24.0 24.1 "Adaptive value of sex in microbial pathogens". Infect. Genet. Evol. 8 (3): 267–85. 2008. doi:10.1016/j.meegid.2008.01.002. PMID 18295550.
External links
- Proteobacteria information from Palaeos.
- Proteobacteria. – J. P. Euzéby: List of Prokaryotic names with Standing in Nomenclature.
Wikidata ☰ Q130999 entry
