Biology:Translocase
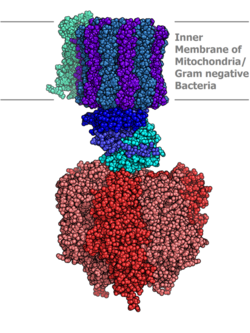
Translocase is a general term for a protein that assists in moving another molecule, usually across a cell membrane. These enzymes catalyze the movement of ions or molecules across membranes or their separation within membranes. The reaction is designated as a transfer from “side 1” to “side 2” because the designations “in” and “out”, which had previously been used, can be ambiguous.[1] Translocases are the most common secretion system in Gram positive bacteria.
It is also a historical term for the protein now called elongation factor G, due to its function in moving the transfer RNA (tRNA) and messenger RNA (mRNA) through the ribosome.
History
The enzyme classification and nomenclature list was first approved by the International Union of Biochemistry in 1961. Six enzyme classes had been recognized based on the type of chemical reaction catalyzed, including oxidoreductases (EC 1), transferases (EC 2), hydrolases (EC 3), lyases (EC 4), isomerases (EC 5) and ligases (EC 6). However, it became apparent that none of these could describe the important group of enzymes that catalyse the movement of ions or molecules across membranes or their separation within membranes. Several of these involve the hydrolysis of ATP and had been previously classified as ATPases (EC 3.6.3.-), although the hydrolytic reaction is not their primary function. In August 2018, the International Union of Biochemistry and Molecular Biology classified these enzymes under a new enzyme class (EC) of translocases (EC 7).[2]
Mechanism of catalysis
The reaction most translocases catalyse is:
- AX + Bside 1|| = A + X + || Bside 2[3]
A clear example of an enzyme that follows this scheme is H+-transporting two-sector ATPase:
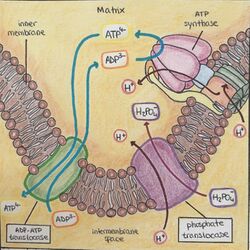
- ATP + H2O + 4 H+side 1 = ADP + phosphate + 4 H+side 2This ATPase carries out the dephosphorylation of ATP into ADP while it transports H+ to the other side of the membrane.[4]
However, other enzymes that also fall into this category do not follow the same reaction scheme. This is the case of ascorbate ferrireductase:
- ascorbateside 1 + Fe(III)side 2 = monodehydroascorbateside 1 + Fe(II)side 2
In which the enzyme only transports an electron in the catalysation of an oxidoreductase reaction between a molecule and an inorganic cation located on different sides of the membrane.[5]
Function
The basic function, as already mentioned (see: Translocase § Definition), is to "catalyse the movement of ions or molecules across membranes or their separation within membranes". This form of membrane transport is classified under active membrane transport, an energy-requiring process of pumping molecules and ions across membranes against a concentration gradient.[6]
Translocases biological importance relies primarily on their critical function, in the way that they provide movement across the cell's membrane in many cellular processes that are substantial, such as:
- Oxidative phosphorylation
- ADP/ATP translocase (ANT) imports adenosine diphosphate ADP from the cytosol and exports ATP from the mitochondrial matrix, which are key transport steps for oxidative phosphorylation in eukaryotic organisms. ADP from the cytosol is transported back into the mitochondrion for ATP synthesis and the synthesised ATP, produced from oxidative phosphorylation, is exported out of the mitochondrion for use in the cytosol, providing the cells with its main energy currency.[7]
- Protein import into mitochondria
- Hundreds of proteins encoded by the nucleus are required for mitochondrial metabolism, growth, division, and partitioning to daughter cells, and all of these proteins must be imported into the organelle.[8] Translocase of the outer membrane (TOM) and translocase of the inner membrane (TIM) mediate the import of proteins into the mitochondrion. The translocase of the outer membrane (TOM) sorts proteins via several mechanisms either directly to the outer membrane, the intermembrane space, or the translocase of the inner membrane (TIM). Then, generally, the TIM23 machinery mediates protein translocation into the matrix and the TIM22 machinery mediates insertion into the inner membrane.[9]
- Fatty acids import into mitochondria (Carnitine Shuttle System)
- Carnitine-acylcarnitine translocase (CACT) catalyzes both unidirectional transport of carnitine and carnitine/acylcarnitine exchange in the inner mitochondrial membrane, allowing the import of long-chain fatty acids into the mitochondria where they are oxidized by the β-oxidation pathway.[10] The mitochondrial membrane is impermeable to long-chain fatty acids, hence the need for this translocation.[11]
Classification
The enzyme subclasses designate the types of components that are being transferred, and the sub-subclasses indicate the reaction processes that provide the driving force for the translocation.[12]
EC 7.1 Catalysing the translocation of hydrons[13]
This subclass contains translocases that catalyze the translocation of hydrons.[14] Based on the reaction they are linked to, EC 7.1 can be further classified into:
- EC 7.1.1 Hydron translocation or charge separation linked to oxidoreductase reactions
- EC 7.1.2 Hydron translocation linked to the hydrolysis of a nucleoside triphosphate
- EC 7.1.3 Hydron translocation linked to the hydrolysis of diphosphate
An important translocase contained in this group is ATP synthase, also known as EC 7.1.2.2.
EC 7.2 Catalysing the translocation of inorganic cations and their chelates
This subclass contains translocases that transfer inorganic cations (metal cations).[15] Based on the reaction they're linked to, EC 7.2 can be further classified into:
- EC 7.2.1 Translocation of inorganic cations linked to oxidoreductase reactions
- EC 7.2.2 Translocation of inorganic cations linked to the hydrolysis of a nucleoside triphosphate
- EC 7.2.4 Translocation of inorganic cations linked to decarboxylation
An important translocase contained in this group is Na+/K+ pump, also known as EC 7.2.2.13.
EC 7.3 Catalysing the translocation of inorganic anions
This subclass contains translocases that transfer inorganic cations anions. Subclasses are based on the reaction processes that provide the driving force for the translocation. At present only one subclass is represented: EC 7.3.2 Translocation of inorganic anions linked to the hydrolysis of a nucleoside triphosphate.[16]
- 7.3.2.1 ABC-type phosphate transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the high affinity uptake of phosphate anions. Unlike P-type ATPases, it does not undergo phosphorylation during the transport process.[17]
- ATP + H2O + phosphate [phosphate - binding protein][side 1] = ADP + phosphate + phosphate [side 2] + [phosphate - binding protein][side 1]
- 7.3.2.2 ABC-type phosphonate transporter
- The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of phosphonate and organophosphate anions.[18]
- ATP + H2O + phosphonate [phosphonate-binding protein][side 1] = ADP + phosphate + phosphonate [side 2] + [phosphonate- binding protein][side 1]
- 7.3.2.3 ABC-type sulfate transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme from Escherichia coli can interact with either of two periplasmic binding proteins and mediates the high affinity uptake of sulfate and thiosulfate. May also be involved in the uptake of selenite, selenate and possibly molybdate. Does not undergo phosphorylation during the transport.[19]
- ATP + H2O + sulfate [sulfate - binding protein] [side 1] = ADP + phosphate + sulfate [side 2] + [sulfate - binding protein][side 1]
- 7.3.2.4 ABC-type nitrate transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of nitrate, nitrite, and cyanate.[20]
- ATP + H2O + nitrate [nitrate - binding protein][side 1] = ADP + phosphate + nitrate [side 2] + [nitrate - binding protein][side 1]
- 7.3.2.5 ABC-type molybdate transporter
- The expected taxonomic range for this enzyme is: Archaea, Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the high-affinity import of molybdate and tungstate. Does not undergo phosphorylation during the transport process.[21]
- ATP + H2O + molybdate [molybdate - binding protein][side 1] = ADP + phosphate + molybdate [side 2] + [molybdate - binding protein][side 1]
- 7.3.2.6 ABC-type tungstate transporter
- The expected taxonomic range for this enzyme is: Archaea, Bacteria. The enzyme, characterized from the archaeon Pyrococcus furiosus, the Gram-positive bacterium Eubacterium acidaminophilum and the Gram-negative bacterium Campylobacter jejuni, interacts with an extracytoplasmic substrate binding protein and mediates the import of tungstate into the cell for incorporation into tungsten-dependent enzymes.[22]
- ATP + H2O + tungstate [tungstate - binding protein][side 1] = ADP + phosphate + tungstate [side 2] + [tungstate - binding protein][side 1]
EC 7.4 Catalysing the translocation of amino acids and peptides
Subclasses are based on the reaction processes that provide the driving force for the translocation. At present there is only one subclass: EC 7.4.2 Translocation of amino acids and peptides linked to the hydrolysis of a nucleoside triphosphate.[23]
- 7.4.2.1 ABC-type polar-amino-acid transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein and mediates the import of polar amino acids. This entry comprises bacterial enzymes that import Histidine, Arginine, Lysine, Glutamine, Glutamate, Aspartate, ornithine, octopine and nopaline.[24]
- ATP + H2O + polar amino acid [polar amino acid-binding protein][side 1] = ADP + phosphate + polar amino acid [side 2] + [polar amino acid-binding protein][side1]
- 7.4.2.2 ABC-type nonpolar-amino-acid transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. The enzyme, found in bacteria, interacts with an extracytoplasmic substrate binding protein. This entry comprises enzymes that import Leucine, Isoleucie and Valine.[25]
- ATP + H2O + non polar amino acid [non polar amino acid - binding protein][side 1] = ADP + phosphate + non polar amino acid [side 2] + [non polar amino acid - binding protein][side 1]
- 7.4.2.3 ABC-type mitochondrial protein-transporting ATPase
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. A non-phosphorylated, non-ABC (ATP-binding cassette) ATPase involved in the transport of proteins or preproteins into mitochondria using the TIM (Translocase of the Inner Membrane) protein complex. TIM is the protein transport machinery of the mitochondrial inner membrane that contains three essential TIM proteins: Tim17 and Tim23 are thought to build a preprotein translocation channel while Tim44 interacts transiently with the matrix heat-shock protein Hsp70 to form an ATP-driven import motor.[26]
- ATP + H2O + mitochondrial protein [side 1] = ADP + phosphate + mitochondrial protein [side 2]
- 7.4.2.4 ABC-type chloroplast protein-transporting ATPase
- The enzyme appears in viruses and cellular organisms. Involved in the transport of proteins or preproteins into chloroplast stroma (several ATPases may participate in this process).[27]
- ATP + H2O + chloroplast protein [side 1] = ADP + phosphate + chloroplast protein [side 2]
- 7.4.2.5 ABC-type protein transporter
- The expected taxonomic range for this enzyme is: Eukaryota, Bacteria. This entry stands for a family of bacterial enzymes that are dedicated to the secretion of one or several closely related proteins belonging to the toxin, protease and lipase families. Examples from Gram-negative bacteria include α-hemolysin, cyclolysin, colicin V and siderophores, while examples from Gram-positive bacteria include bacteriocin, subtilin, competence factor and pediocin.[28]
- ATP + H2O + protein [side 1] = ADP + phosphate + protein [side 2]
- 7.4.2.6 ABC-type oligopeptide transporter
- A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the import of oligopeptides of varying nature. The binding protein determines the specificity of the system. Does not undergo phosphorylation during the transport process.[29]
- ATP + H2O + oligopeptide [oligopeptide - binding protein][side 1] = ADP + phosphate + oligopeptide [side 2] + [oligopeptide - binding protein][side 1]
- 7.4.2.7 ABC-type alpha-factor-pheromone transporter
- The enzyme appears in viruses and cellular organisms characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. Does not undergo phosphorylation during the transport process. A yeast enzyme that exports the α-factor sex pheromone.[30]
- ATP + H2O + alpha factor [side 1] = ADP + phosphate + alpha factor [side 2]
- 7.4.2.8 ABC-type protein-secreting ATPase
- The expected taxonomic range for this enzyme is: Archaea, Bacteria. A non-phosphorylated, non-ABC (ATP-binding cassette) ATPase that is involved in protein transport.[31]
- ATP + H2O + cellular protein [side 1] = ADP + phosphate + cellular protein [side 2]
- 7.4.2.9 ABC-type dipeptide transporter
- The enzyme appears in viruses and cellular organisms. ATP-binding cassette (ABC) type transporter, characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the uptake of dipeptides and tripeptides.[32]
- ATP + H2O + dipeptide [dipeptide - binding protein][side 1] = ADP + phosphate + [side 2] + [dipeptide - binding protein][side 1]
- 7.4.2.10 ABC-type glutathione transporter
- A prokaryotic ATP-binding cassette (ABC) type transporter, characterized by the presence of two similar ATP-binding domains/proteins and two integral membrane domains/proteins. The enzyme from the bacterium Escherichia coli is a heterotrimeric complex that interacts with an extracytoplasmic substrate binding protein to mediate the uptake of glutathione.[33]
- ATP + H2O glutathione [glutathione - binding protein][side 1] = ADP + phosphate + glutathione [side 2] + [glutathione - binding protein][side 1]
- 7.4.2.11 ABC-type methionine transporter
- A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and functions to import methionine.[34][35]
- (1) ATP + H2O + L-methionine [methionine - binding protein][side 1] = ADP + phosphate + L-methionine [side 2] + [methionine - binding protein][side 1]
- (2) ATP + H2O + D-methionine [methionine - binding protein][side 1] = ADP + phosphate + D-methionine [side 2] + [methionine - binding protein][side 1]
- 7.4.2.12 ABC-type cystine transporter
- A bacterial enzyme that interacts with an extracytoplasmic substrate binding protein and mediates the high affinity import of trace cystine. The enzyme from Escherichia coli K-12 can import both isomers of cystine and a variety of related molecules including djenkolate, lanthionine, diaminopimelate and homocystine.[36]
- (1) ATP + H2O + L-cystine [cystine - binding protein][side 1] = ADP + phosphate + L-cystine [side 2] + [cystine - binding protein][side 1]
- (2) ATP + H2O + D-cystine [cystine - binding protein][side 1] = ADP + phosphate + D-cystine [side 2] + [cystine - binding protein][side 1]
EC 7.5 Catalysing the translocation of carbohydrates and their derivatives
- EC 7.5.2 Linked to the hydrolysis of a nucleoside triphosphate
EC 7.6 Catalysing the translocation of other compounds
- EC 7.6.2 Linked to the hydrolysis of a nucleoside triphosphate
Examples
- ornithine translocase (SLC25A15), associated with ornithine translocase deficiency.
- carnitine-acylcarnitine translocase (SLC25A20), associated with carnitine-acylcarnitine translocase deficiency.
- Translocase of outer mitochondrial membrane 40 (TOMM40), a protein encoded by the TOMM40 gene, whose alleles differentially impact the risk for Alzheimer's disease
References
- ↑ "EC class 7". https://www.enzyme-database.org/cinfo.php?c=7&sc=0&ssc=.
- ↑ "Translocases (EC 7): A new EC Class". https://www.enzyme-database.org/news.php.
- ↑ "A Brief Guide to Enzyme Nomenclature and Classification". 2018. https://iubmb.org/wp-content/uploads/sites/2790/2018/11/A-Brief-Guide-to-Enzyme-Classification-and-Nomenclature-rev.pdf.
- ↑ "ExplorEnz: EC 7.1.2.2". https://www.enzyme-database.org/query.php?ec=7.1.2.2.
- ↑ "BRENDA - Information on EC 7.2.1.3 - ascorbate ferrireductase (transmembrane)". https://www.brenda-enzymes.org/enzyme.php?ecno=7.2.1.3.
- ↑ "Active Transport" (in en). CK-12 Foundation. https://www.ck12.org/biology/active-transport/lesson/Active-Transport-Advanced-BIO-ADV/.
- ↑ "The transport mechanism of the mitochondrial ADP/ATP carrier". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. Channels and transporters in cell metabolism 1863 (10): 2379–93. October 2016. doi:10.1016/j.bbamcr.2016.03.015. PMID 27001633.
- ↑ "Protein translocation across mitochondrial membranes: what a long, strange trip it is". Cell 83 (4): 517–9. November 1995. doi:10.1016/0092-8674(95)90089-6. PMID 7585952.
- ↑ "Protein translocation pathways of the mitochondrion". FEBS Letters. Birmingham Issue 476 (1–2): 27–31. June 2000. doi:10.1016/S0014-5793(00)01664-1. PMID 10878244.
- ↑ "Diseases caused by defects of mitochondrial carriers: a review". Biochimica et Biophysica Acta (BBA) - Bioenergetics. 15th European Bioenergetics Conference 2008 1777 (7–8): 564–78. 2008-07-01. doi:10.1016/j.bbabio.2008.03.008. PMID 18406340.
- ↑ "Fatty Acids -- Transport and Regeneration". https://library.med.utah.edu/NetBiochem/FattyAcids/8_4.html.
- ↑ "EC class 7". https://www.enzyme-database.org/cinfo.php?c=7&sc=0&ssc=.
- ↑ Hydron is a generic term that includes protons (1H+), deuterons (2H+) and tritons (3H+).
- ↑ "EC 7.1 - Catalysing the translocation of hydrons". https://www.ebi.ac.uk/intenz/query?cmd=SearchEC&ec=7.1.
- ↑ "EC 7.2 - Catalysing the translocation of inorganic cations". https://www.ebi.ac.uk/intenz/query?cmd=SearchEC&ec=7.2.
- ↑ "IntEnz - EC 7.3". https://www.ebi.ac.uk/intenz/query?cmd=SearchEC&ec=7.3.
- ↑ "BRENDA - Information on EC 7.3.2.1 - ABC-type phosphate transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.3.2.1.
- ↑ "BRENDA - Information on EC 7.3.2.2 - ABC-type phosphonate transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.3.2.2.
- ↑ "BRENDA - Information on EC 7.3.2.3 - ABC-type sulfate transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.3.2.3.
- ↑ "BRENDA - Information on EC 7.3.2.4 - ABC-type nitrate transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.3.2.4.
- ↑ "BRENDA - Information on EC 7.3.2.5 - ABC-type molybdate transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.3.2.5.
- ↑ "BRENDA - Information on EC 7.3.2.6 - ABC-type tungstate transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.3.2.6.
- ↑ "IntEnz - EC 7.4". https://www.ebi.ac.uk/intenz/query?cmd=SearchEC&ec=7.4.
- ↑ "BRENDA - Information on EC 7.4.2.1 - ABC-type polar-amino-acid transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.4.2.1.
- ↑ "BRENDA - Information on EC 7.4.2.2 - ABC-type nonpolar-amino-acid transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.4.2.2.
- ↑ "BRENDA - Information on EC 7.4.2.3 - mitochondrial protein-transporting ATPase". https://www.brenda-enzymes.org/enzyme.php?ecno=7.4.2.3.
- ↑ "BRENDA - Information on EC 7.4.2.4 - chloroplast protein-transporting ATPase". https://www.brenda-enzymes.org/enzyme.php?ecno=7.4.2.4.
- ↑ "BRENDA - Information on EC 7.4.2.5 - ABC-type protein transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.4.2.5.
- ↑ "BRENDA - Information on EC 7.4.2.6 - ABC-type oligopeptide transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.4.2.6.
- ↑ "BRENDA - Information on EC 7.4.2.7 - ABC-type alpha-factor-pheromone transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.4.2.7.
- ↑ "BRENDA - Information on EC 7.4.2.8 - protein-secreting ATPase". https://www.brenda-enzymes.org/enzyme.php?ecno=7.4.2.8.
- ↑ "BRENDA - Information on EC 7.4.2.9 - ABC-type dipeptide transporter". https://www.brenda-enzymes.org/enzyme.php?ecno=7.4.2.9.
- ↑ "Rhea - Annotated reactions database". https://www.rhea-db.org/reaction?id=29791.
- ↑ "Rhea - Annotated reactions database". https://www.rhea-db.org/reaction?id=29779.
- ↑ "Rhea - Annotated reactions database". https://www.rhea-db.org/reaction?id=29767.
- ↑ "IntEnz - EC 7.4.2.12". https://www.ebi.ac.uk/intenz/query?cmd=SearchEC&ec=7.4.2.12.
 |