Biology:Glycogen phosphorylase
| Phosphorylase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
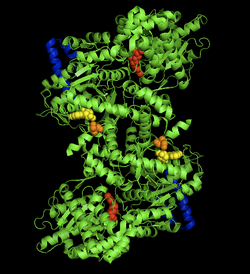 The crystal structure of the rabbit muscle glycogen phosphorylase-AMP complex. AMP allosteric site (yellow), phosphorylated Ser14 (orange), glycogen binding site (blue), catalytic site (red).[1] | |||||||||
| Identifiers | |||||||||
| EC number | 2.4.1.1 | ||||||||
| CAS number | 9035-74-9 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
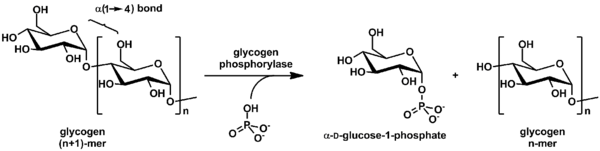
Glycogen phosphorylase is one of the phosphorylase enzymes (EC 2.4.1.1). Glycogen phosphorylase catalyzes the rate-limiting step in glycogenolysis in animals by releasing glucose-1-phosphate from the terminal alpha-1,4-glycosidic bond. Glycogen phosphorylase is also studied as a model protein regulated by both reversible phosphorylation and allosteric effects.
Mechanism
Glycogen phosphorylase breaks up glycogen into glucose subunits (see also figure below):
(α-1,4 glycogen chain)n + Pi ⇌ (α-1,4 glycogen chain)n-1 + α-D-glucose-1-phosphate.[2]
Glycogen is left with one fewer glucose molecule, and the free glucose molecule is in the form of glucose-1-phosphate. In order to be used for metabolism, it must be converted to glucose-6-phosphate by the enzyme phosphoglucomutase.
Although the reaction is reversible in vitro, within the cell the enzyme only works in the forward direction as shown below because the concentration of inorganic phosphate is much higher than that of glucose-1-phosphate.[2]
Glycogen phosphorylase can act only on linear chains of glycogen (α1-4 glycosidic linkage). Its work will immediately come to a halt four residues away from α1-6 branch (which are exceedingly common in glycogen). In these situations, the debranching enzyme is necessary, which will straighten out the chain in that area. In addition, the enzyme transferase shifts a block of 3 glucosyl residues from the outer branch to the other end, and then a α1-6 glucosidase enzyme is required to break the remaining (single glucose) α1-6 residue that remains in the new linear chain. After all this is done, glycogen phosphorylase can continue. The enzyme is specific to α1-4 chains, as the molecule contains a 30-angstrom-long crevice with the same radius as the helix formed by the glycogen chain; this accommodates 4-5 glucosyl residues, but is too narrow for branches. This crevice connects the glycogen storage site to the active, catalytic site.
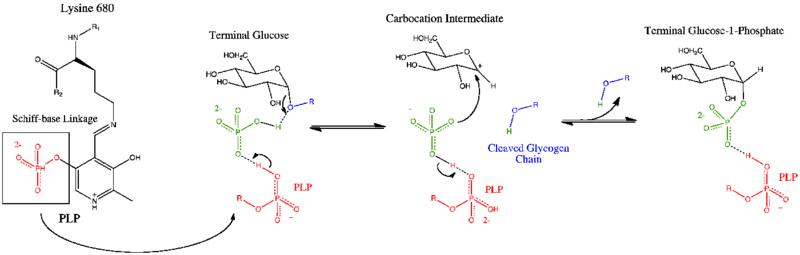
Glycogen phosphorylase has a pyridoxal phosphate (PLP, derived from Vitamin B6) at each catalytic site. Pyridoxal phosphate links with basic residues (in this case Lys680) and covalently forms a Schiff base. Once the Schiff base linkage is formed, holding the PLP molecule in the active site, the phosphate group on the PLP readily donates a proton to an inorganic phosphate molecule, allowing the inorganic phosphate to in turn be deprotonated by the oxygen forming the α-1,4 glycosidic linkage. PLP is readily deprotonated because its negative charge is not only stabilized within the phosphate group, but also in the pyridine ring, thus the conjugate base resulting from the deprotonation of PLP is quite stable. The protonated oxygen now represents a good leaving group, and the glycogen chain is separated from the terminal glycogen in an SN1 fashion, resulting in the formation of a glucose molecule with a secondary carbocation at the 1 position. Finally, the deprotonated inorganic phosphate acts as a nucleophile and bonds with the carbocation, resulting in the formation of glucose-1-phosphate and a glycogen chain shortened by one glucose molecule.
There is also an alternative proposed mechanism involving a positively charged oxygen in a half-chair conformation.[3]
Structure
The glycogen phosphorylase monomer is a large protein, composed of 842 amino acids with a mass of 97.434 kDa in muscle cells. While the enzyme can exist as an inactive monomer or tetramer, it is biologically active as a dimer of two identical subunits.[4]
In mammals, the major isozymes of glycogen phosphorylase are found in muscle, liver, and brain. The brain type is predominant in adult brain and embryonic tissues, whereas the liver and muscle types are predominant in adult liver and skeletal muscle, respectively.[5]
The glycogen phosphorylase dimer has many regions of biological significance, including catalytic sites, glycogen binding sites, allosteric sites, and a reversibly phosphorylated serine residue. First, the catalytic sites are relatively buried, 15Å from the surface of the protein and from the subunit interface.[6] This lack of easy access of the catalytic site to the surface is significant in that it makes the protein activity highly susceptible to regulation, as small allosteric effects could greatly increase the relative access of glycogen to the site.
Perhaps the most important regulatory site is Ser14, the site of reversible phosphorylation very close to the subunit interface. The structural change associated with phosphorylation, and with the conversion of phosphorylase b to phosphorylase a, is the arrangement of the originally disordered residues 10 to 22 into α helices. This change increases phosphorylase activity up to 25% even in the absence of AMP, and enhances AMP activation further.[7]
The allosteric site of AMP binding on muscle isoforms of glycogen phosphorylase are close to the subunit interface just like Ser14. Binding of AMP at this site, corresponding in a change from the T state of the enzyme to the R state, results in small changes in tertiary structure at the subunit interface leading to large changes in quaternary structure.[8] AMP binding rotates the tower helices (residues 262-278) of the two subunits 50˚ relative to one another through greater organization and intersubunit interactions. This rotation of the tower helices leads to a rotation of the two subunits by 10˚ relative to one another, and more importantly disorders residues 282-286 (the 280s loop) that block access to the catalytic site in the T state but do not in the R state.[6]
The final, perhaps most curious site on the glycogen phosphorylase protein is the so-called glycogen storage site. Residues 397-437 form this structure, which allows the protein to covalently bind to the glycogen chain a full 30 Å from the catalytic site . This site is most likely the site at which the enzyme binds to glycogen granules before initiating cleavage of terminal glucose molecules. In fact, 70% of dimeric phosphorylase in the cell exists as bound to glycogen granules rather than free floating.[9]
Clinical significance
| phosphorylase, glycogen; muscle (McArdle syndrome, glycogen storage disease type V) | |
|---|---|
| Identifiers | |
| Symbol | PYGM |
| NCBI gene | 5837 |
| HGNC | 9726 |
| OMIM | 608455 |
| RefSeq | NM_005609 |
| UniProt | P11217 |
| Other data | |
| EC number | 2.4.1.1 |
| Locus | Chr. 11 q12-q13.2 |
| phosphorylase, glycogen; liver (Hers disease, glycogen storage disease type VI) | |
|---|---|
| Identifiers | |
| Symbol | PYGL |
| NCBI gene | 5836 |
| HGNC | 9725 |
| OMIM | 232700 |
| RefSeq | NM_002863 |
| UniProt | P06737 |
| Other data | |
| EC number | 2.4.1.1 |
| Locus | Chr. 14 q11.2-24.3 |
| phosphorylase, glycogen; brain | |
|---|---|
| Identifiers | |
| Symbol | PYGB |
| NCBI gene | 5834 |
| HGNC | 9723 |
| OMIM | 138550 |
| RefSeq | NM_002862 |
| UniProt | P11216 |
| Other data | |
| EC number | 2.4.1.1 |
| Locus | Chr. 20 p11.2-p11.1 |
The inhibition of glycogen phosphorylase has been proposed as one method for treating type 2 diabetes.[10] Since glucose production in the liver has been shown to increase in type 2 diabetes patients,[11] inhibiting the release of glucose from the liver's glycogen's supplies appears to be a valid approach. The cloning of the human liver glycogen phosphorylase (HLGP) revealed a new allosteric binding site near the subunit interface that is not present in the rabbit muscle glycogen phosphorylase (RMGP) normally used in studies. This site was not sensitive to the same inhibitors as those at the AMP allosteric site,[12] and most success has been had synthesizing new inhibitors that mimic the structure of glucose, since glucose-6-phosphate is a known inhibitor of HLGP and stabilizes the less active T-state.[13] These glucose derivatives have had some success in inhibiting HLGP, with predicted Ki values as low as 0.016 mM.[14]
Mutations in the muscle isoform of glycogen phosphorylase (PYGM) are associated with glycogen storage disease type V (GSD V, McArdle's Disease). More than 65 mutations in the PYGM gene that lead to McArdle disease have been identified to date.[15][16] Symptoms of McArdle disease include muscle weakness, myalgia, and lack of endurance, all stemming from low glucose levels in muscle tissue.[17]
Mutations in the liver isoform of glycogen phosphorylase (PYGL) are associated with Hers' Disease (glycogen storage disease type VI).[18][19] Hers' disease is often associated with mild symptoms normally limited to hypoglycemia, and is sometimes difficult to diagnose due to residual enzyme activity.[20]
The brain isoform of glycogen phosphorylase (PYGB) has been proposed as a biomarker for gastric cancer.[21]
Regulation
Glycogen phosphorylase is regulated through allosteric control and through phosphorylation. Phosphorylase a and phosphorylase b each exist in two forms: a T (tense) inactive state and an R (relaxed) state. Phosphorylase b is normally in the T state, inactive due to the physiological presence of ATP and Glucose 6 phosphate, and Phosphorylase a is normally in the R state (active). An isoenzyme of glycogen phosphorylase exists in the liver sensitive to glucose concentration, as the liver acts as a glucose exporter. In essence, liver phosphorylase is responsive to glucose, which causes a very responsive transition from the R to T form, inactivating it; furthermore, liver phosphorylase is insensitive to AMP.
Hormones such as epinephrine, insulin and glucagon regulate glycogen phosphorylase using second messenger amplification systems linked to G proteins. Glucagon activates adenylate cyclase through a G protein-coupled receptor (GPCR) coupled to Gs which in turn activates adenylate cyclase to increase intracellular concentrations of cAMP. cAMP binds to and activates protein kinase A (PKA). PKA phosphorylates phosphorylase kinase, which in turn phosphorylates glycogen phosphorylase b at Ser14, converting it into the active glycogen phosphorylase a.
In the liver, glucagon also activates another GPCR that triggers a different cascade, resulting in the activation of phospholipase C (PLC). PLC indirectly causes the release of calcium from the hepatocytes' endoplasmic reticulum into the cytosol. The increased calcium availability binds to the calmodulin subunit and activates glycogen phosphorylase kinase. Glycogen phosphorylase kinase activates glycogen phosphorylase in the same manner mentioned previously.
Glycogen phosphorylase b is not always inactive in muscle, as it can be activated allosterically by AMP.[6][9] An increase in AMP concentration, which occurs during strenuous exercise, signals energy demand. AMP activates glycogen phosphorylase b by changing its conformation from a tense to a relaxed form. This relaxed form has similar enzymatic properties as the phosphorylated enzyme. An increase in ATP concentration opposes this activation by displacing AMP from the nucleotide binding site, indicating sufficient energy stores.
Upon eating a meal, there is a release of insulin, signaling glucose availability in the blood. Insulin indirectly activates protein phosphatase 1 (PP1) and phosphodiesterase via a signal transduction cascade. PP1 dephosphorylates glycogen phosphorylase a, reforming the inactive glycogen phosphorylase b. The phosphodiesterase converts cAMP to AMP. Together, they decrease the concentration of cAMP and inhibit PKA. As a result, PKA can no longer initiate the phosphorylation cascade that ends with formation of (active) glycogen phosphorylase a. Overall, insulin signaling decreases glycogenolysis to preserve glycogen stores in the cell and triggers glycogenesis.[22]
Historical significance
Glycogen phosphorylase was the first allosteric enzyme to be discovered.[8] It was isolated and its activity characterized in detail by Carl F. Cori, Gerhard Schmidt and Gerty T. Cory.[23][24] Arda Green and Gerty Cori crystallized it for the first time in 1943[25] and illustrated that glycogen phosphorylase existed in either the a or b forms depending on its phosphorylation state, as well as in the R or T states based on the presence of AMP.[26]
See also
- AMP deaminase deficiency (MADD)
- Glycogenolysis
- McArdle disease (GSD-V)
- Metabolic myopathies
- Purine nucleotide cycle § Pathology
References
- ↑ PDB: 3E3N
- ↑ 2.0 2.1 "Pyridoxal 5'-phosphate as a catalytic and conformational cofactor of muscle glycogen phosphorylase B". Biochemistry. Biokhimiia 67 (10): 1089–98. October 2002. doi:10.1023/A:1020978825802. PMID 12460107.
- ↑ "The role of pyridoxal 5'-phosphate in glycogen phosphorylase catalysis". Biochemistry 29 (5): 1099–107. February 1990. doi:10.1021/bi00457a001. PMID 2182117.
- ↑ "Phosphorylase: a biological transducer". Trends in Biochemical Sciences 17 (2): 66–71. February 1992. doi:10.1016/0968-0004(92)90504-3. PMID 1566331.
- ↑ "Quantitation of muscle glycogen phosphorylase mRNA and enzyme amounts in adult rat tissues". Biochimica et Biophysica Acta (BBA) - General Subjects 880 (1): 78–90. January 1986. doi:10.1016/0304-4165(86)90122-4. PMID 3510670.
- ↑ 6.0 6.1 6.2 "Glycogen phosphorylase: control by phosphorylation and allosteric effectors". FASEB Journal 6 (6): 2274–82. March 1992. doi:10.1096/fasebj.6.6.1544539. PMID 1544539.
- ↑ "The family of glycogen phosphorylases: structure and function". Critical Reviews in Biochemistry and Molecular Biology 24 (1): 69–99. 1989. doi:10.3109/10409238909082552. PMID 2667896.
- ↑ 8.0 8.1 "Glycogen phosphorylase. The structural basis of the allosteric response and comparison with other allosteric proteins". The Journal of Biological Chemistry 265 (5): 2409–12. February 1990. doi:10.1016/S0021-9258(19)39810-2. PMID 2137445.
- ↑ 9.0 9.1 "Control of phosphorylase activity in a muscle glycogen particle. I. Isolation and characterization of the protein-glycogen complex". The Journal of Biological Chemistry 245 (24): 6642–8. December 1970. doi:10.1016/S0021-9258(18)62582-7. PMID 4320610.
- ↑ "Glucose analog inhibitors of glycogen phosphorylases as potential antidiabetic agents: recent developments". Current Pharmaceutical Design 9 (15): 1177–89. 2003. doi:10.2174/1381612033454919. PMID 12769745.
- ↑ "New drug targets for type 2 diabetes and the metabolic syndrome". Nature 414 (6865): 821–7. December 2001. doi:10.1038/414821a. PMID 11742415. Bibcode: 2001Natur.414..821M.
- ↑ "An engineered liver glycogen phosphorylase with AMP allosteric activation". The Journal of Biological Chemistry 266 (24): 16113–9. August 1991. doi:10.1016/S0021-9258(18)98523-6. PMID 1874749.
- ↑ "The design of potential antidiabetic drugs: experimental investigation of a number of beta-D-glucose analogue inhibitors of glycogen phosphorylase". European Journal of Drug Metabolism and Pharmacokinetics 19 (3): 185–92. Jul 1994. doi:10.1007/BF03188920. PMID 7867660.
- ↑ "Prediction of Ligand−Receptor Binding Free Energy by 4D-QSAR Analysis: Application to a Set of Glucose Analogue Inhibitors of Glycogen Phosphorylase". Journal of Chemical Information and Computer Sciences 39 (6): 1141–1150. Sep 1999. doi:10.1021/ci9900332.
- ↑ "Molecular genetics of McArdle's disease". Current Neurology and Neuroscience Reports 7 (1): 84–92. January 2007. doi:10.1007/s11910-007-0026-2. PMID 17217859.
- ↑ "McArdle disease: molecular genetic update". Acta Myologica 26 (1): 53–7. July 2007. PMID 17915571.
- ↑ "Acute renal failure in McArdle's disease. Report of two cases". The New England Journal of Medicine 286 (23): 1237–41. June 1972. doi:10.1056/NEJM197206082862304. PMID 4502558.
- ↑ "Mutations in the liver glycogen phosphorylase gene (PYGL) underlying glycogenosis type VI". American Journal of Human Genetics 62 (4): 785–91. April 1998. doi:10.1086/301790. PMID 9529348.
- ↑ "Identification of a mutation in liver glycogen phosphorylase in glycogen storage disease type VI". Human Molecular Genetics 7 (5): 865–70. May 1998. doi:10.1093/hmg/7.5.865. PMID 9536091.
- ↑ "A novel mutation (G233D) in the glycogen phosphorylase gene in a patient with hepatic glycogen storage disease and residual enzyme activity". Molecular Genetics and Metabolism 79 (2): 142–5. June 2003. doi:10.1016/S1096-7192(03)00068-4. PMID 12809646.
- ↑ "Gastric and intestinal phenotypes of gastric carcinoma with reference to expression of brain (fetal)-type glycogen phosphorylase". Journal of Gastroenterology 36 (7): 457–64. July 2001. doi:10.1007/s005350170068. PMID 11480789.
- ↑ "The protein phosphatases involved in cellular regulation. Evidence that dephosphorylation of glycogen phosphorylase and glycogen synthase in the glycogen and microsomal fractions of rat liver are catalysed by the same enzyme: protein phosphatase-1". European Journal of Biochemistry 156 (1): 101–10. April 1986. doi:10.1111/j.1432-1033.1986.tb09554.x. PMID 3007140.
- ↑ "The Synthesis of a Polysaccharide From Glucose-1-Phosphate in Muscle Extract". Science 89 (2316): 464–5. May 1939. doi:10.1126/science.89.2316.464. PMID 17731092. Bibcode: 1939Sci....89..464C. https://www.science.org/doi/10.1126/science.89.2316.464.
- ↑ "The kinetics of the enzymatic synthesis of glycogen from glucose-1-phosphate". Journal of Biological Chemistry 135 (2): 733–756. July 1940. doi:10.1016/S0021-9258(18)73136-0. https://www.jbc.org/content/135/2/733.short.
- ↑ "Crystalline Muscle Phosphorylase I. Preparation, Properties, and Molecular Weight". Journal of Biological Chemistry 151: 21–29. July 7, 1943. doi:10.1016/S0021-9258(18)72110-8. https://www.jbc.org/content/151/1/21.short.
- ↑ "Crystalline muscle phosphorylase II prosthetic group". Journal of Biological Chemistry 151 (1): 21–29. July 1943. doi:10.1016/S0021-9258(18)72113-3.
Further reading
- "Chapter 17: Glycogen Metabolism". Biochemistry (2nd ed.). New York: J. Wiley & Sons. 1995. ISBN 978-0-471-58651-7. https://archive.org/details/biochemistry00voet_0.
- "Chapter 18: Glycogen Metabolism". Biochemistry (3rd ed.). New York: J. Wiley & Sons. 2004. ISBN 978-0-471-19350-0. https://archive.org/details/biochemistry00voet_1.
- "Glycogen Phosphorylase". Molecule of the Month. RCSB Protein Data Bank. 2001-12-01. http://www.rcsb.org/pdb/static.do?p=education_discussion%2Fmolecule_of_the_month%2Fpdb24_1.html.
- "Glycogen Metabolism". Molecular Biochemistry I. Rensselaer Polytechnic Institute. http://rpi.edu/dept/bcbp/molbiochem/MBWeb/mb1/part2/glycogen.htm#animat1.
External links
- GeneReviews/NCBI/NIH/UW entry on Glycogen Storage Disease Type VI - Hers disease
- Glycogen+phosphorylase at the US National Library of Medicine Medical Subject Headings (MeSH)
- Overview of all the structural information available in the PDB for UniProt: P11217 (Human muscle Glycogen phosphorylase) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: P06737 (Human liver Glycogen phosphorylase) at the PDBe-KB.
- Overview of all the structural information available in the PDB for UniProt: P11216 (Human brain Glycogen phosphorylase) at the PDBe-KB.
 |