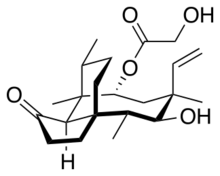
Chemistry:Pleuromutilin
| |
| |
| Names | |
|---|---|
| Preferred IUPAC name
(3aS,4R,5S,6S,8R,9R,9aR,10R)-6-Ethenyl-5-hydroxy-4,6,9,10-tetramethyl-1-oxodecahydro-3a,9-propanocyclopenta[8]annulen-8-yl hydroxyacetate | |
| Identifiers | |
3D model (JSmol)
|
|
| ChEMBL | |
| ChemSpider | |
| KEGG | |
PubChem CID
|
|
| UNII | |
| |
| |
| Properties | |
| C22H34O5 | |
| Molar mass | 378.509 g/mol |
| Melting point | 170-171 °C |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Pleuromutilin and its derivatives are antibacterial drugs that inhibit protein synthesis in bacteria by binding to the peptidyl transferase component of the 50S subunit of ribosomes.[1][2]
This class of antibiotics includes the licensed drugs lefamulin (for systemic use in humans), retapamulin (approved for topical use in humans), valnemulin and tiamulin (approved for use in animals) and the investigational drug azamulin.[citation needed]
History
Pleuromutilin was discovered as an antibiotic in 1951.[3][4] It is derived from the fungi Omphalina mutila (formerly Pleurotus mutilus) and Clitopilus passeckerianus (formerly Pleurotus passeckerianus),[3] and has also been found in Drosophila subatrata, Clitopilus scyphoides, and some other Clitopilus species.[5]
Total synthesis
The total synthesis of pleuromutilin has been reported.[6][7][8][9]
Biosynthesis
Pleuromutilin belongs to the class of secondary metabolites known as terpenes, which are produced in fungi through the mevalonate pathway (MEP pathway).[10] Its synthetic bottleneck lays on the production of the precursor GGPP and the following formation of the tricyclic structure, which is catalyzed by Pl-cyc, a bifunctional diterpene synthase (DTS). This Cyclase shows a new class II DTS activity, catalyzes a ring contraction and the formation of a 5-6-bicyclic ring structure. Specifically, DTS shows two catalytic distinguishable domains: On the one hand it has at the N-terminal region a class II DTS domain, which catalyzes a cascade cyclization, resulting in a decalin core. Subsequently, variable 1,2-proton and methyl shifts occur to translocate the carbocation towards one of the two interconnecting C-atoms and this intermediate induces a base-catalyzed ring contraction. Therefore, class II DTS promotes in general a ring contraction during the cyclisation of GGPP. On the other hand, at the C-terminal end it has a class I DTS domain, which catalyzes a conjugated dephosphorylation, generating the 8-membered cyclic core, followed by a 1,5-proton shift and a stereospecific hydroxylation to obtain premutilin.[11]
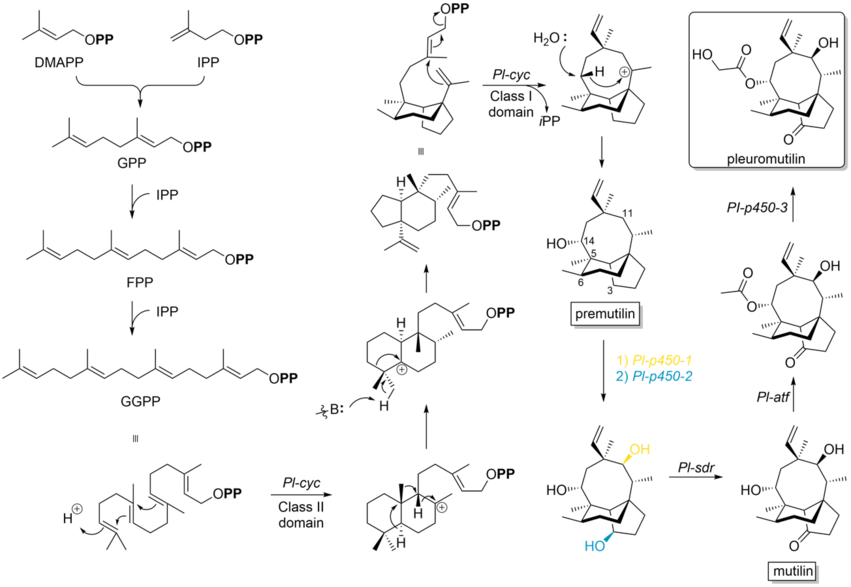
Additionally, three cytochrome P450s (Pl-p450-1, Pl-p450-2 and Pl-p450-3) are involved in the final steps of the pleuromutilin biosynthetic pathway.[14] The P450-1 and P450-2 are essential for hydroxylation of two ring structures regarding the premutilin skeleton, oxidating specifically at position C-11 and C-3, respectively. The short-chain dehydrogenase/reductase enzyme (Pl-sdr) has a regiospecific activity and converts the 3-hydroxy group to a ketone, forming the intermediate mutilin. Acetyltransferase (Pl-atf) catalyzes the transfer of acetyl group to 14-OH of mutilin. Finally, Pl-p450-3 hydroxylates the α-methyl group of the acetyl side chain generating pleuromutilin.[14][15]
References
- ↑ Maffioli, Sonia Ilaria (2013). "A chemist's survey of different antibiotic classes". in Gualerzi, Claudio O.; Brandi, Letizia; Fabbretti, Attilio et al.. Antibiotics: Targets, Mechanisms and Resistance. Wiley-VCH. pp. 1–22. doi:10.1002/9783527659685.ch1. ISBN 978-3-527-65968-5.
- ↑ "A novel pleuromutilin antibacterial compound, its binding mode and selectivity mechanism". Sci Rep 6(39004): 39004. Dec 2016. doi:10.1038/srep39004. PMID 27958389. Bibcode: 2016NatSR...639004E.
- ↑ 3.0 3.1 "Antibiotic substances from basidiomycetes: VIII. Pleurotus multilus (Fr.) Sacc. and Pleurotus passeckerianus Pilat.". Proceedings of the National Academy of Sciences of the United States of America 37 (9): 570–574. September 1951. doi:10.1073/pnas.37.9.570. PMID 16589015. Bibcode: 1951PNAS...37..570K.
- ↑ "The pleuromutilin antibiotics: a new class for human use". Current Opinion in Investigational Drugs 11 (2): 182–91. February 2010. PMID 20112168.
- ↑ "Establishing molecular tools for genetic manipulation of the pleuromutilin-producing fungus Clitopilus passeckerianus". Applied and Environmental Microbiology 75 (22): 7196–204. November 2009. doi:10.1128/AEM.01151-09. PMID 19767458. Bibcode: 2009ApEnM..75.7196K.
- ↑ Gibbons, E. Grant (1982). "Total synthesis of (±)-pleuromutilin". Journal of the American Chemical Society 104 (6): 1767–1769. doi:10.1021/ja00370a067.
- ↑ Boeckman, Robert K.; Springer, Dane M.; Alessi, Thomas R. (1989). "Synthetic studies directed toward naturally occurring cyclooctanoids. 2. A stereocontrolled assembly of (±)-pleuromutilin via a remarkable sterically demanding oxy-Cope rearrangement". Journal of the American Chemical Society 111 (21): 8284–8286. doi:10.1021/ja00203a043.
- ↑ "Total synthesis of (+)-pleuromutilin". Chemistry: A European Journal 19 (21): 6718–23. May 2013. doi:10.1002/chem.201300968. PMID 23589420.
- ↑ "A modular and enantioselective synthesis of the pleuromutilin antibiotics". Science 356 (6341): 956–959. June 2017. doi:10.1126/science.aan0003. PMID 28572392. Bibcode: 2017Sci...356..956M.
- ↑ "Isoprenoid biosynthesis: the evolution of two ancient and distinct pathways across genomes". Proceedings of the National Academy of Sciences of the United States of America 97 (24): 13172–7. November 2000. doi:10.1073/pnas.240454797. PMID 11078528. Bibcode: 2000PNAS...9713172M.
- ↑ 11.0 11.1 "Biosynthesis of bioactive natural products from Basidiomycota". Organic & Biomolecular Chemistry 17 (5): 1027–1036. January 2019. doi:10.1039/C8OB02774A. PMID 30608100.
- ↑ Birch, A.J.; Holzapfel, C.W.; Rickards, R.W. (January 1966). "The structure and some aspects of the biosynthesis of pleuromutilin" (in en). Tetrahedron 22: 359–387. doi:10.1016/S0040-4020(01)90949-4. https://linkinghub.elsevier.com/retrieve/pii/S0040402001909494.
- ↑ "Some studies in the biosynthesis of terpenes and related compounds". Pure and Applied Chemistry 17 (3): 331–48. 1968-01-01. doi:10.1351/pac196817030331. PMID 5729285.
- ↑ 14.0 14.1 "Heterologous expression reveals the biosynthesis of the antibiotic pleuromutilin and generates bioactive semi-synthetic derivatives". Nature Communications 8 (1): 1831. November 2017. doi:10.1038/s41467-017-01659-1. PMID 29184068. Bibcode: 2017NatCo...8.1831A.
- ↑ "Identification and manipulation of the pleuromutilin gene cluster from Clitopilus passeckerianus for increased rapid antibiotic production". Scientific Reports 6 (1): 25202. May 2016. doi:10.1038/srep25202. PMID 27143514. Bibcode: 2016NatSR...625202B.
Further reading
- "Interaction of pleuromutilin derivatives with the ribosomal peptidyl transferase center". Antimicrobial Agents and Chemotherapy 50 (4): 1458–62. April 2006. doi:10.1128/AAC.50.4.1458-1462.2006. PMID 16569865.
- "A click chemistry approach to pleuromutilin conjugates with nucleosides or acyclic nucleoside derivatives and their binding to the bacterial ribosome". Journal of Medicinal Chemistry 51 (16): 4957–67. August 2008. doi:10.1021/jm800261u. PMID 18680270.
 |

