Biology:Iodothyronine deiodinase
| Type I thyroxine 5'-deiodinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.21.99.4 | ||||||||
| CAS number | 70712-46-8 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Type II thyroxine 5-deiodinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 1.21.99.3 | ||||||||
| CAS number | 74506-30-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Type III thyroxine 5-deiodinase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Mouse iodothyronine deiodinase 3 catalytic core rendered from PDB entry 4TR3 [1] | |||||||||
| Identifiers | |||||||||
| EC number | 1.97.1.11 | ||||||||
| CAS number | 74506-30-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Iodothyronine deiodinases (EC 1.21.99.4 and EC 1.21.99.3) are a subfamily of deiodinase enzymes important in the activation and deactivation of thyroid hormones. Thyroxine (T4), the precursor of 3,5,3'-triiodothyronine (T3) is transformed into T3 by deiodinase activity. T3, through binding a nuclear thyroid hormone receptor, influences the expression of genes in practically every vertebrate cell.[2][3] Iodothyronine deiodinases are unusual in that these enzymes contain selenium, in the form of an otherwise rare amino acid selenocysteine.[4][5][6]
These enzymes are not to be confused with the iodotyrosine deiodinases that are also deiodinases, but not members of the iodothyronine family. The iodotyrosine deiodinases (unlike the iodothyronine deiodinases) do not use selenocysteine or selenium. The iodotyrosine enzymes work on iodinated single tyrosine residue molecules to scavenge iodine, and do not use as substrates the double-tyrosine residue molecules of the various iodothyronines.
Activation and inactivation
In tissues, deiodinases can either activate or inactivate thyroid hormones:
- Activation occurs by conversion of thyroxine (T4) to the active hormone triiodothyronine (T3) through the removal of an iodine atom on the outer ring.
- Inactivation of thyroid hormones occurs by removal of an iodine atom on the inner ring, which converts thyroxine to the inactive reverse triiodothyronine (rT3), or which converts the active triiodothyronine to diiodothyronine (T2).
The major part of thyroxine deiodination occurs within the cells.
Deiodinase 2 activity can be regulated by ubiquitination:
- The covalent attachment of ubiquitin inactivates D2 by disrupting dimerization and targets it to degradation in the proteosome.[7]
- Deubiquitination removing ubiquitin from D2 restores its activity and prevents proteosomal degradation.[7]
- The Hedgehog cascade acts to increase D2 ubiquitination through WSB1 activity, decreasing D2 activity.[7][8]
D-propranolol inhibits thyroxine deiodinase, thereby blocking the conversion of T4 to T3, providing some though minimal therapeutic effect.[citation needed]
Reactions
| 400px|left|Reactions catalyzed by specific deiodinase isoforms | 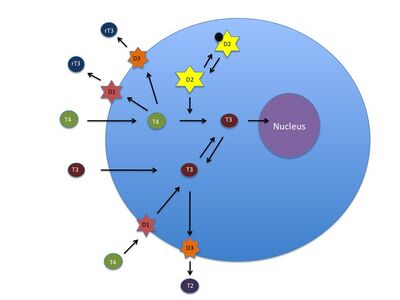 |
Structure
The three deiodinase enzymes share certain structural features in common although their sequence identity is lower than 50%. Each enzyme weighs between 29 and 33kDa.[7] Deiodinases are dimeric integral membrane proteins with single transmembrane segments and large globular heads (see below).[9] They share a TRX fold that contains the active site including the rare selenocysteine amino acid and two histidine residues.[7][10] Selenocysteine is coded by a UGA codon, which generally signifies termination of a peptide through a stop codon. In point mutation experiments with Deiodinase 1 changing UGA to the stop codon TAA resulted in a complete loss of function, while changing UGA to cysteine (TGT) caused the enzyme to operate at around 10% normal efficiency.[11] In order for UGA to be read as a selenocysteine amino acid instead of a stop codon, it is necessary that a downstream stem loop sequence, the selenocysteine insertion sequence (SECIS), be present to bind with SECIS binding protein-2 (SBP-2), which binds with elongation factor EFsec.[7] The translation of selenocysteine is not efficient,[12] even though it is important to the functioning of the enzyme. Deiodinase 2 is localized to the ER membrane while Deiodinase 1 and 3 are found in the plasma membrane.[7]
The related catalytic domains of Deiodinases 1-3 feature a thioredoxine-related peroxiredoxin fold.[13] The enzymes catalyze a reductive elimination of iodine, thereby oxidizing themselves similar to Prx, followed by a reductive recycling of the enzyme.
Types
| Type I iodothyronine deiodinase | |
|---|---|
| Identifiers | |
| Symbol | DIO1 |
| Alt. symbols | TXDI1 |
| NCBI gene | 1733 |
| HGNC | 2883 |
| OMIM | 147892 |
| RefSeq | NM_000792 |
| UniProt | P49895 |
| Other data | |
| EC number | 1.21.99.3 |
| Locus | Chr. 1 p32-p33 |
| Type II iodothyronine deiodinase | |
|---|---|
| Identifiers | |
| Symbol | DIO2 |
| Alt. symbols | TXDI2, SelY |
| NCBI gene | 1734 |
| HGNC | 2884 |
| OMIM | 601413 |
| RefSeq | NM_000793 |
| UniProt | Q92813 |
| Other data | |
| EC number | 1.21.99.4 |
| Locus | Chr. 14 q24.2-24.3 |
| Type III iodothyronine deiodinase | |
|---|---|
| Identifiers | |
| Symbol | DIO3 |
| Alt. symbols | TXDI3 |
| NCBI gene | 1735 |
| HGNC | 2885 |
| OMIM | 601038 |
| PDB | 4TR3 |
| RefSeq | NM_001362 |
| UniProt | P55073 |
| Other data | |
| EC number | 1.97.1.11 |
| Locus | Chr. 14 q32 |
In most vertebrates, there are three types of enzymes that can deiodinate thyroid hormones:
| Type | Location | Function |
| type I (DI) | is commonly found in the liver and kidney | DI can deiodinate both rings[14] |
| type II deiodinase (DII) | is found in the heart, skeletal muscle, CNS, fat, thyroid, and pituitary[15] | DII can only deiodinate the outer ring of the prohormone thyroxine and is the major activating enzyme (the already inactive reverse triiodothyronine is also degraded further by DII) |
| type III deiodinase (DIII) | found in the fetal tissue and the placenta; also present throughout the brain, except in the pituitary[16] | DIII can only deiodinate the inner ring of thyroxine or triiodothyronine and is the major inactivating enzyme |
Function
Deiodinase 1 both activates T4 to produce T3 and inactivates T4. Besides its increased function in producing extrathyroid T3 in patients with hyperthyroidism, its function is less well understood than D2 or D3 [2][7] Deiodinase 2, located in the ER membrane, converts T4 into T3 and is a major source of the cytoplasmic T3 pool.[2] Deiodinase 3 prevents T4 activation and inactivates T3.[9] D2 and D3 are important in homeostatic regulation in maintaining T3 levels at the plasma and cellular levels. In hyperthyroidism D2 is down regulated and D3 is upregulated to clear extra T3, while in hypothyroidism D2 is upregulated and D3 is downregulated to increase cytoplasmic T3 levels.[2][7]
Serum T3 levels remain fairly constant in healthy individuals, but D2 and D3 can regulate tissue specific intracellular levels of T3 to maintain homeostasis since T3 and T4 levels may vary by organ. Deiodinases also provide spatial and temporal developmental control of thyroid hormone levels. D3 levels are highest early in development and decrease over time, while D2 levels are high at moments of significant metamorphic change in tissues. Thus D2 enables production of sufficient T3 at necessary time points while D3 may shield tissue from overexposure to T3.[12]
Also, iodothyronine deiodinases (type 2 y 3; DIO2 and DIO3, respectively) respond to seasonal changes in photoperiod-driven melatonin secretion and govern peri-hypothalamic catabolism of the prohormone thyroxine (T4). In long summer days, the production of hypothalamic T3 increase due to DIO-2-mediated conversion of T4 to the biologically active hormone. This process allows to active anabolic neuroendocrine pathways that maintain reproductive competence and increase body weight. However, during the adaptation to reproductively inhibitory photoperiods, the levels of T3 decrease due to peri-hypothalamic DIO3 expression that catabolizes T4 and T3 into receptor inactive amines.[17][18]
Deiodinase 2 also plays a significant role in thermogenesis in brown adipose tissue (BAT). In response to sympathetic stimulation, dropping temperature, or overfeeding BAT, D2 increases oxidation of fatty acids and uncouples oxidative phosphorylation via uncoupling protein, causing mitochondrial heat production. D2 increases during cold stress in BAT and increases intracellular T3 levels. In D2 deficient models, shivering is a behavioral adaptation to the cold. However, heat production is much less efficient than uncoupling lipid oxidation.[19][20]
Disease relevance
In cardiomyopathy the heart reverts to a fetal gene programming due to the overload of the heart. Like during fetal development, thyroid hormone levels are low in the overloaded heart tissue in a local hypothyroid state, with low levels of deiodinase 1 and deiodinase 2. Although deiodinase 3 levels in a normal heart are generally low, in cardiomyopathy deiodinase 3 activity is increased to decrease energy turnover and oxygen consumption.[7]
Hypothyroidism is a disease diagnosed by decreased levels of serum thyroxine (T4). Presentation in adults leads to decreased metabolism, increased weight gain, and neuropsychiatric complications.[21] During development, hypothyroidism is considered more severe and leads to neurotoxicity as cretinism or other human cognitive disorders,[22] altered metabolism and underdeveloped organs. Medication and environmental exposures can result in hypothyroidism with changes in deiodinase enzyme activity. The drug iopanoic acid (IOP) decreased cutaneous cell proliferation by inhibition of deiodinase enzyme type 1 or 2 reducing the conversion of T4 to T3. The environmental chemical DE-71, a PBDE pentaBDE brominated flame retardant decreased hepatic deiodinase I transcription and enzyme activity in neonatal rats with hypothyroidism.[23]
Quantifying enzyme activity
In vitro, including cell culture experiments, deiodination activity is determined by incubating cells or homogenates with high amounts of labeled thyroxine (T4) and required cosubstrates. As a measure of deiodination, the production of radioactive iodine and other physiological metabolites, in particular T3 or reverse T3, are determined and expressed (e.g. as fmol/mg protein/minute).[24][25]
In vivo, deiodination activity is estimated from equilibrium levels of free T3 and free T4. A simple approximation is T3/T4 ratio,[26] a more elaborate approach is calculating sum activity of peripheral deiodinases (GD) from free T4, free T3 and parameters for protein binding, dissociation and hormone kinetics.[27][28] In atypical cases, this last approach can benefit from measurements of TBG, but usually only requires measurement of TSH, fT3 and fT4, and as such has no added laboratory requirements besides the measurement of the same.
See also
- Iodotyrosine deiodinase
- Selenium, section Evolution in biology
References
- ↑ "Crystal structure of mammalian selenocysteine-dependent iodothyronine deiodinase suggests a peroxiredoxin-like catalytic mechanism". Proceedings of the National Academy of Sciences of the United States of America 111 (29): 10526–31. July 2014. doi:10.1073/pnas.1323873111. PMID 25002520. Bibcode: 2014PNAS..11110526S.
- ↑ 2.0 2.1 2.2 2.3 "Deiodinases: implications of the local control of thyroid hormone action". The Journal of Clinical Investigation 116 (10): 2571–9. October 2006. doi:10.1172/JCI29812. PMID 17016550.
- ↑ "Gene regulation by thyroid hormone". Trends in Endocrinology and Metabolism 11 (6): 207–11. August 2000. doi:10.1016/s1043-2760(00)00263-0. PMID 10878749.
- ↑ "The selenoenzyme family of deiodinase isozymes controls local thyroid hormone availability". Reviews in Endocrine & Metabolic Disorders 1 (1–2): 49–58. January 2000. doi:10.1023/A:1010012419869. PMID 11704992.
- ↑ "Local activation and inactivation of thyroid hormones: the deiodinase family". Molecular and Cellular Endocrinology 151 (1–2): 103–19. May 1999. doi:10.1016/S0303-7207(99)00040-4. PMID 10411325.
- ↑ "The deiodinase family: selenoenzymes regulating thyroid hormone availability and action". Cellular and Molecular Life Sciences 57 (13–14): 1853–63. December 2000. doi:10.1007/PL00000667. PMID 11215512.
- ↑ 7.00 7.01 7.02 7.03 7.04 7.05 7.06 7.07 7.08 7.09 "Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling". Endocrine Reviews 29 (7): 898–938. December 2008. doi:10.1210/er.2008-0019. PMID 18815314.
- ↑ "The Hedgehog-inducible ubiquitin ligase subunit WSB-1 modulates thyroid hormone activation and PTHrP secretion in the developing growth plate". Nature Cell Biology 7 (7): 698–705. July 2005. doi:10.1038/ncb1272. PMID 15965468.
- ↑ 9.0 9.1 Bianco AC. "Thyroid hormone action starts and ends by deiodination". Bianco Lab & The University of Miami. http://deiodination.org/.
- ↑ "Cloning and expression of a 5'-iodothyronine deiodinase from the liver of Fundulus heteroclitus". Endocrinology 138 (2): 642–8. February 1997. doi:10.1210/endo.138.2.4904. PMID 9002998.
- ↑ "Type I iodothyronine deiodinase is a selenocysteine-containing enzyme". Nature 349 (6308): 438–40. January 1991. doi:10.1038/349438a0. PMID 1825132. Bibcode: 1991Natur.349..438B.
- ↑ 12.0 12.1 "The deiodinase family of selenoproteins". Thyroid 7 (4): 655–68. August 1997. doi:10.1089/thy.1997.7.655. PMID 9292958.
- ↑ "Crystal structure of mammalian selenocysteine-dependent iodothyronine deiodinase suggests a peroxiredoxin-like catalytic mechanism". Proceedings of the National Academy of Sciences of the United States of America 111 (29): 10526–31. July 2014. doi:10.1073/pnas.1323873111. PMID 25002520. Bibcode: 2014PNAS..11110526S.
- ↑ "Activation and inactivation of thyroid hormone by type I iodothyronine deiodinase". FEBS Letters 344 (2–3): 143–6. May 1994. doi:10.1016/0014-5793(94)00365-3. PMID 8187873.
- ↑ Holtorf K (2012). "Deiodinases". National Academy of Hypothyroidism. http://nahypothyroidism.org/deiodinases/.
- ↑ "The role of thyroid hormone deiodination in the regulation of hypothalamo-pituitary function". Neuroendocrinology 38 (3): 254–60. March 1984. doi:10.1159/000123900. PMID 6371572.
- ↑ "Genome sequencing and transcriptome analyses of the Siberian hamster hypothalamus identify mechanisms for seasonal energy balance". Proceedings of the National Academy of Sciences of the United States of America 116 (26): 13116–13121. June 2019. doi:10.1073/pnas.1902896116. PMID 31189592. Bibcode: 2019PNAS..11613116B.
- ↑ "Hypothalamic thyroid hormone catabolism acts as a gatekeeper for the seasonal control of body weight and reproduction". Endocrinology 148 (8): 3608–17. August 2007. doi:10.1210/en.2007-0316. PMID 17478556.
- ↑ "Intracellular conversion of thyroxine to triiodothyronine is required for the optimal thermogenic function of brown adipose tissue". The Journal of Clinical Investigation 79 (1): 295–300. January 1987. doi:10.1172/JCI112798. PMID 3793928.
- ↑ "The type 2 iodothyronine deiodinase is essential for adaptive thermogenesis in brown adipose tissue". The Journal of Clinical Investigation 108 (9): 1379–85. November 2001. doi:10.1172/JCI13803. PMID 11696583.
- ↑ "The role of thyroid hormones in depression". European Journal of Endocrinology 138 (1): 1–9. January 1998. doi:10.1530/eje.0.1380001. PMID 9461307.
- ↑ "Role of late maternal thyroid hormones in cerebral cortex development: an experimental model for human prematurity". Cerebral Cortex 20 (6): 1462–75. June 2010. doi:10.1093/cercor/bhp212. PMID 19812240.
- ↑ "Effects of perinatal PBDE exposure on hepatic phase I, phase II, phase III, and deiodinase 1 gene expression involved in thyroid hormone metabolism in male rat pups". Toxicological Sciences 107 (1): 27–39. January 2009. doi:10.1093/toxsci/kfn230. PMID 18978342.
- ↑ "Type 2 iodothyronine deiodinase in rat pituitary tumor cells is inactivated in proteasomes". The Journal of Clinical Investigation 102 (11): 1895–9. December 1998. doi:10.1172/JCI4672. PMID 9835613.
- ↑ "Hypoxia-inducible factor induces local thyroid hormone inactivation during hypoxic-ischemic disease in rats". The Journal of Clinical Investigation 118 (3): 975–83. March 2008. doi:10.1172/JCI32824. PMID 18259611.
- ↑ "The serum triiodothyronine to thyroxine (T3/T4) ratio in various thyroid disorders and after Levothyroxine replacement therapy". Hormones 3 (2): 120–6. 2004. doi:10.14310/horm.2002.11120. PMID 16982586.
- ↑ Dietrich JW (2002). Der Hypophysen-Schilddrüsen-Regelkreis. Berlin, Germany: Logos-Verlag Berlin. 3897228505. ISBN 978-3-89722-850-4. OCLC 50451543.
- ↑ "Influence of low protein diet on nonthyroidal illness syndrome in chronic renal failure". Endocrine 27 (3): 283–8. August 2005. doi:10.1385/ENDO:27:3:283. PMID 16230785.
Further reading
- (in de) Biochemie und Pathobiochemie (Springer-Lehrbuch) (German ed.). Berlin: Springer. 2006. pp. 847–861. ISBN 978-3-540-32680-9.
External links
- Deiodinase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |

