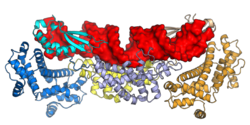
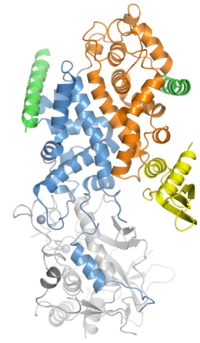
Biology:Ribonuclease III
| Ribonuclease III domain | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
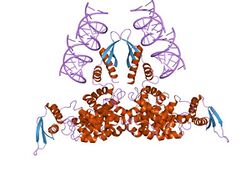 Ribonuclease III structure interacting with double stranded RNA. | |||||||||||
| Identifiers | |||||||||||
| Symbol | RNase_III | ||||||||||
| Pfam | PF00636 | ||||||||||
| InterPro | IPR000999 | ||||||||||
| PROSITE | PDOC00448 | ||||||||||
| SCOP2 | 1jfz / SCOPe / SUPFAM | ||||||||||
| |||||||||||
Ribonuclease III (RNase III or RNase C)[1](BRENDA 3.1.26.3) is a type of ribonuclease that recognizes dsRNA and cleaves it at specific targeted locations to transform them into mature RNAs.[2] These enzymes are a group of endoribonucleases that are characterized by their ribonuclease domain, which is labelled the RNase III domain.[3] They are ubiquitous compounds in the cell and play a major role in pathways such as RNA precursor synthesis, RNA Silencing, and the pnp autoregulatory mechanism.[4][5]
Types of RNase III
The RNase III superfamily is divided into four known classes: 1, 2, 3, and 4. Each class is defined by its domain structure.[6]
Class 1 RNase III
- Class 1 RNase III enzymes have a homodimeric structure whose function is to cleave dsRNA into multiple subunits. It is a Mg2+-dependent endonuclease and is largely found in bacteria and bacteriophage. Class 1 RNase III have been found in Glomeromycotan fungi, which was suspected to be the result of horizontal gene transfer from cyanobacteria.[7] Among the RNases III in the class are the rnc from E. coli. Typically, class I enzymes possess a single RNase III domain (RIIID) followed by a dsRNA-binding domain (dsRBD).[6] They process precursors to ribosomal RNA (rRNA), small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA). The basic dsRNA cleavage function of Class 1 RNase III is retained in most of the organisms in which it is present. However, in a number of species the function has changed and taken on different or additional biological roles.[8]
Class 2 RNase III
- Class II is defined by the presence of an N-terminal domain (NTD), a RIIID, and a dsRBD. Class II is found in some fungi species.[6] They process precursors to rRNA, snRNA, and snoRNA.
- Yeast nucleases with the Class 2 RNase III domain:[11]
Class 3 RNase III
- Class 3 RNases III include the Drosha family of enzymes known to function in maturation of precursors to microRNA (miRNA).[14]
Class 4 RNase III
- Class 4 RNases III include the Dicer family of enzymes known to function in RNA interference (RNAi).[15] Class 4 III RNases are S-RNase components. It is a component of the self-incompatibility system in Rosaceae, Solanaceae, and Plantaginaceae. They are recruited to cope with various environmental stress scenarios.[16]
- Dicer enzymes process dsRNA substrates into small RNA fragments of individual size ranging from 21-27 nucleotides in length.[17] Dicer has an N-terminal helicase/ATPase domain which is followed by another domain of an unknown function. It also comprises the centrally positioned PAZ domain and a C-terminal configuration which includes one dsRBD and two RNase III catalytic domains.[18] Interactions of Dicer occurs with other proteins, which includes TRBP, PACT, and Ago2.[19] RNAs that are produced by Dicer act as guides for a sequence of particular silencing of cognate genes through RNAi and related pathways.[17]
Human proteins containing RNase III domain
See also
- RncO
References
- ↑ Filippov, Valery; Solovyev, Victor; Filippova, Maria; Gill, Sarjeet S. (7 March 2000). "A novel type of RNase III family proteins in eukaryotes". Gene 245 (1): 213–221. doi:10.1016/S0378-1119(99)00571-5. PMID 10713462.
- ↑ Zamore, Phillip D. (December 2001). "Thirty-Three Years Later, a Glimpse at the Ribonuclease III Active Site". Molecular Cell 8 (6): 1158–1160. doi:10.1016/S1097-2765(01)00418-X. PMID 11885596.
- ↑ Conrad, Christian; Rauhut, Reinhard (February 2002). "Ribonuclease III: new sense from nuisance". The International Journal of Biochemistry & Cell Biology 34 (2): 116–129. doi:10.1016/S1357-2725(01)00112-1. PMID 11809414.
- ↑ Inada, T.; Nakamura, Y. (1995). "Lethal double-stranded RNA processing activity of ribonuclease III in the absence of SuhB protein of Escherichia coli". Biochimie 77 (4): 294–302. doi:10.1016/0300-9084(96)88139-9. PMID 8589060.
- ↑ Park, Hongmarn; Yakhnin, Helen; Connolly, Michael; Romeo, Tony; Babitzke, Paul; Gourse, R. L. (15 December 2015). "CsrA Participates in a PNPase Autoregulatory Mechanism by Selectively Repressing Translation of Transcripts That Have Been Previously Processed by RNase III and PNPase". Journal of Bacteriology 197 (24): 3751–3759. doi:10.1128/JB.00721-15. PMID 26438818.
- ↑ Jump up to: 6.0 6.1 6.2 Liang Y-H, Lavoie M, Comeau M-A, Elela SA, Ji X. Structure of a Eukaryotic RNase III Post-Cleavage Complex Reveals a Double- Ruler Mechanism for Substrate Selection. Molecular cell. 2014;54(3):431-444. doi:10.1016/j.molcel.2014.03.006.
- ↑ Soon-Jae Lee, Mengxuan Kong, Paul Harrison, Mohamed Hijri; Conserved proteins of the RNA interference system in the arbuscular mycorrhizal fungus Rhizoglomus irregulare provide new insight into the evolutionary history of Glomeromycota, Genome Biology and Evolution, , evy002, https://doi.org/10.1093/gbe/evy002
- ↑ Kreuze, Jan F.; Savenkov, Eugene I.; Cuellar, Wilmer; Li, Xiangdong; Valkonen, Jari P. T. (1 June 2005). "Viral Class 1 RNase III Involved in Suppression of RNA Silencing" (in en). Journal of Virology 79 (11): 7227–7238. doi:10.1128/JVI.79.11.7227-7238.2005. ISSN 0022-538X. PMID 15890961.
- ↑ "rnc - Ribonuclease 3 - Escherichia coli (strain K12) - rnc gene & protein". UniProt Consortium. https://www.uniprot.org/uniprot/P0A7Y0.
- ↑ Glow, D.; Pianka, D.; Sulej, A. A.; Kozlowski, Lukasz P.; Czarnecka, J.; Chojnowski, G.; Skowronek, K. J.; Bujnicki, J. M. (2015). "Sequence-specific cleavage of dsRNA by Mini-III RNase". Nucleic Acids Research 43 (5): 2864–2873. doi:10.1093/nar/gkv009. ISSN 0305-1048. PMID 25634891.
- ↑ Wu, Chang-Xian; Xu, Xian-Jin; Zheng, Ke; Liu, Fang; Yang, Xu-Dong; Chen, Chuang-Fu; Chen, Huan-Chun; Liu, Zheng-Fei (1 April 2016). "Characterization of ribonuclease III from Brucella". Gene 579 (2): 183–192. doi:10.1016/j.gene.2015.12.068. PMID 26778206.
- ↑ "RNT1/YMR239C Overview". Stanford University. https://www.yeastgenome.org/locus/S000004852.
- ↑ "pac1 (SPBC119.11c)". EMBL-EBI. http://www.pombase.org/spombe/result/SPBC119.11c.
- ↑ "A novel type of RNase III family proteins in eukaryotes". Gene 245 (1): 213–221. Mar 2000. doi:10.1016/S0378-1119(99)00571-5. PMID 10713462.
- ↑ "Role for a bidentate ribonuclease in the initiation step of RNA interference". Nature 409 (6818): 363–6. 2001. doi:10.1038/35053110. PMID 11201747.

- ↑ Rojas, Hernán; Floyd, Brice; Morriss, Stephanie C.; Bassham, Diane; MacIntosh, Gustavo C.; Goldraij, Ariel (1 July 2015). "NnSR1, a class III non-S-RNase specifically induced in Nicotiana alata under phosphate deficiency, is localized in endoplasmic reticulum compartments". Plant Science 236: 250–259. doi:10.1016/j.plantsci.2015.04.012. PMID 26025538.
- ↑ Jump up to: 17.0 17.1 MacRae, Ian J; Doudna, Jennifer A (February 2007). "Ribonuclease revisited: structural insights into ribonuclease III family enzymes". Current Opinion in Structural Biology 17 (1): 138–145. doi:10.1016/j.sbi.2006.12.002. PMID 17194582.
- ↑ Redko, Yulia; Bechhofer, David H.; Condon, Ciarán (June 2008). "Mini-III, an unusual member of the RNase III family of enzymes, catalyses 23S ribosomal RNA maturation in B. subtilis". Molecular Microbiology 68 (5): 1096–1106. doi:10.1111/j.1365-2958.2008.06207.x. PMID 18363798.
- ↑ Nicholson, Allen W. (January 2014). "Ribonuclease III mechanisms of double-stranded RNA cleavage". Wiley Interdisciplinary Reviews: RNA 5 (1): 31–48. doi:10.1002/wrna.1195. PMID 24124076.
- ↑ "Tissue expression of DICER1 - Summary". The Human Protein Atlas. http://www.proteinatlas.org/ENSG00000100697-DICER1/tissue.
- ↑ "Tissue expression of DROSHA - Summary". The Human Protein Atlas. http://www.proteinatlas.org/ENSG00000113360-DROSHA/tissue.
External links
- RNase+III at the US National Library of Medicine Medical Subject Headings (MeSH)
- EC 3.1.26.3
 |