Biology:Cytomegalovirus
| Cytomegalovirus | |
|---|---|
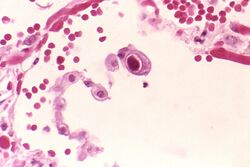
| |
| Typical "owl eye" intranuclear inclusion indicating CMV infection of a lung pneumocyte[1] | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Duplodnaviria |
| Kingdom: | Heunggongvirae |
| Phylum: | Peploviricota |
| Class: | Herviviricetes |
| Order: | Herpesvirales |
| Family: | Orthoherpesviridae |
| Subfamily: | Betaherpesvirinae |
| Genus: | Cytomegalovirus |
| Species | |
|
See text | |
| Synonyms[2] | |
| |
Cytomegalovirus (CMV) (from cyto- 'cell' via Greek κύτος kútos- 'container' + μέγας mégas 'big, megalo-' + -virus via Latin vīrus 'poison') is a genus of viruses in the order Herpesvirales, in the family Herpesviridae,[3] in the subfamily Betaherpesvirinae. Humans and other primates serve as natural hosts. The 11 species in this genus include human betaherpesvirus 5 (HCMV, human cytomegalovirus, HHV-5), which is the species that infects humans. Diseases associated with HHV-5 include mononucleosis and pneumonia,[4][5] and congenital CMV in infants can lead to deafness and ambulatory problems.[6]
In the medical literature, most mentions of CMV without further specification refer implicitly to human CMV. Human CMV is the most studied of all cytomegaloviruses.[7]
MX2/MXB was identified as a restriction factor for herpesviruses, which acts at a very early stage of the replication cycle and MX2/MXB restriction of herpesvirus requires GTPase activity.[8]
Taxonomy
Within the Herpesviridae, CMV belongs to the Betaherpesvirinae subfamily, which also includes the genera Muromegalovirus and Roseolovirus (human herpesvirus 6 and human betaherpesvirus 7).[9] It is also related to other herpesviruses within the Alphaherpesvirinae subfamily, which includes herpes simplex viruses 1 and 2 and varicella-zoster virus, and the Gammaherpesvirinae subfamily, which includes Epstein–Barr virus and Kaposi's sarcoma-associated herpesvirus.[7]
Several species of Cytomegalovirus have been identified and classified for different mammals.[9] The most studied is Human cytomegalovirus (HCMV), which is also known as Human betaherpesvirus 5 (HHV-5). Other primate CMV species include Chimpanzee cytomegalovirus (CCMV) that infects chimpanzees and orangutans, and Simian cytomegalovirus (SCCMV) and Rhesus cytomegalovirus (RhCMV) that infect macaques; CCMV is known as both Panine beta herpesvirus 2 (PaHV-2) and Pongine betaherpesvirus 4 (PoHV-4).[10] SCCMV is called cercopithecine betaherpesvirus 5 (CeHV-5)[11] and RhCMV, Cercopithecine betaherpesvirus 8 (CeHV-8).[12] A further two viruses found in the night monkey are tentatively placed in the genus Cytomegalovirus, and are called Herpesvirus aotus 1 and Herpesvirus aotus 3. Rodents also have viruses previously called cytomegaloviruses that are now reclassified under the genus Muromegalovirus; this genus contains Mouse cytomegalovirus (MCMV) is also known as Murid betaherpesvirus 1 (MuHV-1) and the closely related Murid betaherpesvirus 2 (MuHV-2) that is found in rats.[13]
Species
The following 11 species are assigned to the genus in ICTV 2022:[5]
- Cytomegalovirus aotinebeta1
- Cytomegalovirus cebinebeta1
- Cytomegalovirus cercopithecinebeta5
- Cytomegalovirus humanbeta5
- Cytomegalovirus macacinebeta3
- Cytomegalovirus macacinebeta8
- Cytomegalovirus mandrillinebeta1
- Cytomegalovirus paninebeta2
- Cytomegalovirus papiinebeta3
- Cytomegalovirus papiinebeta4
- Cytomegalovirus saimiriinebeta4
Structure
Viruses in Cytomegalovirus are enveloped, with icosahedral, spherical to pleomorphic, and round geometries, and T=16 symmetry. The diameter is around 150–200 nm. Genomes are linear and nonsegmented, around 200 kb in length.[4]
| Genus | Structure | Symmetry | Capsid | Genomic arrangement | Genomic segmentation |
|---|---|---|---|---|---|
| Cytomegalovirus | Spherical pleomorphic | T=16 | Enveloped | Linear | Monopartite |
Genome
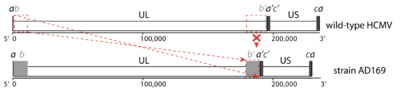
Herpesviruses have some of the largest genomes among human viruses, often encoding hundreds of proteins. For instance, the double‑stranded DNA (dsDNA) genome of wild-type HCMV strains has a size of around 235 kb and encodes at least 208 proteins. It is thus longer than all other human herpesviruses and one of the longest genomes of all human viruses in general. It has the characteristic herpesvirus class E genome architecture, consisting of two unique regions (unique long UL and unique short US), both flanked by a pair of inverted repeats (terminal/internal repeat long TRL/IRL and internal/terminal repeat short IRS/TRS). Both sets of repeats share a region of a few hundred bps, the so-called "a sequence"; the other regions of the repeats are sometimes referred to as "b sequence" and "c sequence".[14]
Life cycle
Viral replication is nuclear and lysogenic. Entry into the host cell is achieved by attachment of the viral glycoproteins to host receptors, which mediates endocytosis. Replication follows the dsDNA bidirectional replication model. DNA templated transcription, with some alternative splicing mechanism is the method of transcription. Translation takes place by leaky scanning. The virus exits the host cell by nuclear egress, and budding. Humans and monkeys serve as the natural hosts. Transmission routes are dependent on coming into contact with bodily fluids (such as saliva, urine, and genital secretions) from an infected individual.[4][15]
| Genus | Host details | Tissue tropism | Entry details | Release details | Replication site | Assembly site | Transmission |
|---|---|---|---|---|---|---|---|
| Cytomegalovirus | humans; monkeys | Epithelial mucosa | Glycoproteins | Budding | Nucleus | Nucleus | Urine; saliva; congenital |
All herpesviruses share a characteristic ability to remain latent within the body over long periods. Although they may be found throughout the body, CMV infections are frequently associated with the salivary glands in humans and other mammals.[9]
Genetic engineering
The CMV promoter is commonly included in vectors used in genetic engineering work conducted in mammalian cells, as it is a strong promoter and drives constitutive expression of genes under its control.[16]
History
Cytomegalovirus was first observed by German pathologist Hugo Ribbert in 1881 when he noticed enlarged cells with enlarged nuclei present in the cells of an infant.[17] Years later, between 1956 and 1957, Thomas Huckle Weller together with Smith and Rowe independently isolated the virus, known thereafter as "cytomegalovirus".[18] In 1990, the first draft of human cytomegalovirus genome was published,[19] the biggest contiguous genome sequenced at that time.[20]
See also
References
- ↑ "Histopathological detection of owl's eye inclusions is still specific for cytomegalovirus in the era of human herpesviruses 6 and 7". Journal of Clinical Pathology 53 (8): 612–4. August 2000. doi:10.1136/jcp.53.8.612. PMID 11002765.
- ↑ "Classification and nomenclature of viruses. Fifth Report of the International Committee on Taxonomy of Viruses.". Arch. Virol.: 107. 1991. https://ictv.global/ictv/proposals/ICTV%205th%20Report.pdf.
- ↑ "Interventions for the management of CMV-associated anterior segment inflammation". The Cochrane Database of Systematic Reviews 2017 (8): CD011908. August 2017. doi:10.1002/14651858.cd011908.pub2. PMID 28838031.
- ↑ 4.0 4.1 4.2 "Viral Zone". ExPASy. http://viralzone.expasy.org/all_by_species/180.html.
- ↑ 5.0 5.1 "Virus Taxonomy: 2022 Release". International Committee on Taxonomy of Viruses (ICTV). March 2023. https://ictv.global/taxonomy.
- ↑ "Often overlooked, a common infection during pregnancy kickstarts a conversation about newborn screening". STAT. 5 April 2023. https://www.statnews.com/2023/04/05/cmv-screening-congenital-cytomegalovirus-hearing-loss.
- ↑ 7.0 7.1 Sherris Medical Microbiology (4th ed.). McGraw Hill. 2004. pp. 556; 566–9. ISBN 978-0-8385-8529-0.
- ↑ "Human MX2/MxB: a Potent Interferon-Induced Postentry Inhibitor of Herpesviruses and HIV-1". Journal of Virology 92 (24). December 2018. doi:10.1128/JVI.00709-18. PMID 30258007.
- ↑ 9.0 9.1 9.2 Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge, UK: Cambridge University Press. 2007. ISBN 978-0-521-82714-0.
- ↑ "Panine betaherpesvirus 2 (Chimpanzee cytomegalovirus)". https://www.uniprot.org/taxonomy/188763.
- ↑ "Simian cytomegalovirus (strain Colburn)". https://www.uniprot.org/taxonomy/50292.
- ↑ "Macacine betaherpesvirus 3 (Rhesus cytomegalovirus)". https://www.uniprot.org/taxonomy/47929.
- ↑ "Murid herpesvirus 1, complete genome". 13 August 2018. http://www.ncbi.nlm.nih.gov/nuccore/NC_004065.1.
- ↑ 14.0 14.1 "Genomic and functional characteristics of human cytomegalovirus revealed by next-generation sequencing". Viruses 6 (3): 1049–1072. March 2014. doi:10.3390/v6031049. PMID 24603756.
- ↑ "Review of cytomegalovirus shedding in bodily fluids and relevance to congenital cytomegalovirus infection". Reviews in Medical Virology 21 (4): 240–255. July 2011. doi:10.1002/rmv.695. PMID 21674676.
- ↑ Morgan, Kendall (3 April 2014). "Plasmids 101: The Promoter Region – Let's Go!". Addgene Blog. http://blog.addgene.org/plasmids-101-the-promoter-region.
- ↑ "Preface". Cytomegaloviruses: Molecular Biology and Immunology. Horizon Scientific Press. 2006. pp. xxiv. ISBN 9781904455028.
- ↑ "Isolation of intranuclear inclusion producing agents from infants with illnesses resembling cytomegalic inclusion disease". Proceedings of the Society for Experimental Biology and Medicine 94 (1): 4–12. January 1957. doi:10.3181/00379727-94-22841. PMID 13400856.
- ↑ "Analysis of the Protein-Coding Content of the Sequence of Human Cytomegalovirus Strain AD169". Cytomegaloviruses. Current Topics in Microbiology and Immunology. 154. Springer Berlin Heidelberg. 1990. pp. 125–69. doi:10.1007/978-3-642-74980-3_6. ISBN 978-3-642-74982-7.
- ↑ "Human cytomegalovirus genomics and transcriptomics through the lens of next-generation sequencing: revision and future challenges". Virus Genes 55 (2): 138–164. April 2019. doi:10.1007/s11262-018-1627-3. PMID 30604286.
External links
| Classification | |
|---|---|
| External resources |
Wikidata ☰ Q6946 entry
 |


