Biology:C2 domain
| C2 domain | |||||||||
|---|---|---|---|---|---|---|---|---|---|
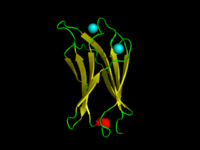 The C2-domain of C.absonum α-toxin (PDB 1OLP). β-strands are shown in yellow, α-helices in red, loops in green. Co-ordinated Calcium ions are in cyan. | |||||||||
| Identifiers | |||||||||
| Symbol | C2 | ||||||||
| Pfam | PF00168 | ||||||||
| InterPro | IPR000008 | ||||||||
| SMART | C2 | ||||||||
| PROSITE | PDOC00380 | ||||||||
| SCOP2 | 1qas / SCOPe / SUPFAM | ||||||||
| OPM superfamily | 45 | ||||||||
| OPM protein | 1ugk | ||||||||
| CDD | cd00030 | ||||||||
| |||||||||
| Phosphoinositide 3-kinase C2 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
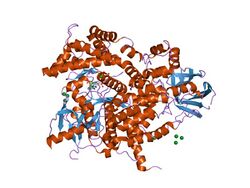 Structure of phosphoinositide 3-kinase.[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | PI3K_C2 | ||||||||||
| Pfam | PF00792 | ||||||||||
| InterPro | IPR002420 | ||||||||||
| SMART | PI3K_C2 | ||||||||||
| PROSITE | PDOC50004 | ||||||||||
| SCOP2 | 1e8x / SCOPe / SUPFAM | ||||||||||
| CDD | cd08380 | ||||||||||
| |||||||||||
A C2 domain is a protein structural domain involved in targeting proteins to cell membranes. The typical version (PKC-C2) has a beta-sandwich composed of 8 β-strands that co-ordinates two or three calcium ions, which bind in a cavity formed by the first and final loops of the domain, on the membrane binding face. Many other C2 domain families don't have calcium binding activity.[2][3]
Coupling with other domains
C2 domains are frequently found coupled to enzymatic domains; for example, the C2 domain in PTEN, brings the phosphatase domain into contact with the plasma membrane, where it can dephosphorylate its substrate, phosphatidylinositol (3,4,5)-trisphosphate (PIP3), without removing it from the membrane - which would be energetically very costly. PTEN consists of two domains, a protein tyrosine phosphatase domain and a C2 domain. This domain pair constitutes a superdomain, a heritable unit that is found in various proteins in fungi, plants and animals.[4] In addition, phosphatidylinositol 3-kinase (PI3-kinase), an enzyme that phosphorylates phosphoinositides on the 3-hydroxyl group of the inositol ring, also uses a C2 domain to bind to the membrane (e.g. 1e8w PDB entry).
Evolution
The C2 domain is currently only known from eukaryotes and the prokaryote Clostridium perfringens where it is part of the alpha-toxin.[5] Over 17 distinct clades of C2 domains have been identified.[2][3] Most C2 families can be traced back to basal eukaryotic species indicating an early diversification before the last eukaryotic common ancestor (LECA). Only the PKC-C2 domain family contains conserved calcium-binding residues, suggesting the typical calcium-dependent membrane interaction is a derived feature limited in PKC-C2 domains.[2]
Calcium and Lipid selectivity
C2 domains are unique among membrane targeting domains in that they show wide range of lipid selectivity for the major components of cell membranes, including phosphatidylserine and phosphatidylcholine. This C2 domain is about 116 amino-acid residues and is located between the two copies of the C1 domain in Protein Kinase C (that bind phorbol esters and diacylglycerol) (see PDOC00379) and the protein kinase catalytic domain (see PDOC00100). Regions with significant homology[6] to the C2-domain have been found in many proteins. The C2 domain is thought to be involved in calcium-dependent phospholipid binding[7] and in membrane targeting processes such as subcellular localisation. Although most C2 domains interact with the membrane (phospholipids) in a Ca2+-dependent manner, some C2 domains can interact with the membrane without binding to Ca2+. Similarly, C2 domains have been evolved to have different specificities for lipids. Many C2 domains such as synaptotagmin C2A, bind to anionic phospholipids (PS or PIP2 containing phospholipids). However, other C2 domains such as cPLA2-α C2 domain bind to zwitterionic lipids (e.g. PC). This diversity and selectivity in Ca2+ and lipid binding suggest that C2 domains are evolved to have different functions.[8]
3D structure
The domain forms an eight-stranded beta sandwich constructed around a conserved 4-stranded motif, designated a C2 key.[9] Calcium binds in a cup-shaped depression formed by the N- and C-terminal loops of the C2-key motif. Structural analyses of several C2 domains have shown them to consist of similar ternary structures in which three Ca2+-binding loops are located at the end of an 8 stranded antiparallel beta sandwich.
Human proteins containing C2 domain
ABR; BAIAP3; BCR; C2CD2; C2CD3; CADPS; CADPS2; CAPN5; CAPN6; CC2D1A; CC2D1B; CPNE1; CPNE2; CPNE3; CPNE4; CPNE5; CPNE6; CPNE7; CPNE8; CPNE9; DAB2IP; DOC2A; DOC2B; DYSF; ESYT1; ESYT3; FAM62A; FAM62B; FAM62C; FER1L3; FER1L5; HECW1; HECW2; ITCH; ITSN1; ITSN2; MCTP1; MCTP2; MTAC2D1; NEDD4; NEDD4L; NEDL1; OTOF; PCLO; PIK3C2A; PIK3C2B; PIK3C2G; PLA2G4A; PLA2G4B; PLA2G4D; PLA2G4E; PLA2G4F; PLCB1; PLCB2; PLCB3; PLCB4; PLCD1; PLCD3; PLCD4; PLCE1; PLCG1; PLCG2; PLCH1; PLCH2; PLCL1; PLCL2; PLCZ1; PRF1; PRKCA; PRKCB1; PRKCE; PRKCG; PRKCH; RAB11FIP1; RAB11FIP2; RAB11FIP5; RASA1; RASA2; RASA3; RASA4; RASAL1; RASAL2; RGS3; RIMS1; RIMS2; RIMS3; RIMS4; RPGRIP1; RPGRIP1L; RPH3A; SGA72M; SMURF1; SMURF2; SYNGAP1; SYT1; SYT10; SYT11; SYT12; SYT13; SYT14; SYT14L; SYT15; SYT16; SYT17; SYT2; SYT3; SYT4; SYT5; SYT6; SYT7; SYT8; SYT9; SYTL1; SYTL2; SYTL3; SYTL4; SYTL5; TOLLIP; UNC13A; UNC13B; UNC13C; UNC13D; WWC2; WWP1; WWP2; PTEN
References
- ↑ "Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine". Molecular Cell 6 (4): 909–19. October 2000. doi:10.1016/S1097-2765(05)00089-4. PMID 11090628.
- ↑ 2.0 2.1 2.2 "Identification of novel families and classification of the C2 domain superfamily elucidate the origin and evolution of membrane targeting activities in eukaryotes". Gene 469 (1–2): 18–30. December 2010. doi:10.1016/j.gene.2010.08.006. PMID 20713135.
- ↑ 3.0 3.1 "Novel transglutaminase-like peptidase and C2 domains elucidate the structure, biogenesis and evolution of the ciliary compartment". Cell Cycle 11 (20): 3861–75. October 2012. doi:10.4161/cc.22068. PMID 22983010.
- ↑ "Superdomains in the protein structure hierarchy: The case of PTP-C2". Protein Science 24 (5): 874–82. May 2015. doi:10.1002/pro.2664. PMID 25694109.
- ↑ Naylor, Claire E.; Eaton, Julian T.; Howells, Angela; Justin, Neil; Moss, David S.; Titball, Richard W.; Basak, Ajit K. (August 1998). "Structure of the key toxin in gas gangrene". Nature Structural & Molecular Biology 5 (8): 738–746. doi:10.1038/1447. ISSN 1545-9993. PMID 9699639.
- ↑ "Mammalian homologues of Caenorhabditis elegans unc-13 gene define novel family of C2-domain proteins". The Journal of Biological Chemistry 270 (42): 25273–80. October 1995. doi:10.1074/jbc.270.42.25273. PMID 7559667.
- ↑ "A single C2 domain from synaptotagmin I is sufficient for high affinity Ca2+/phospholipid binding". The Journal of Biological Chemistry 268 (35): 26386–90. December 1993. PMID 8253763.
- ↑ "C2 domains from different Ca2+ signaling pathways display functional and mechanistic diversity". Biochemistry 40 (10): 3089–100. March 2001. doi:10.1021/bi001968a. PMID 11258923.
- ↑ "Structure of the first C2 domain of synaptotagmin I: a novel Ca2+/phospholipid-binding fold". Cell 80 (6): 929–38. March 1995. doi:10.1016/0092-8674(95)90296-1. PMID 7697723.
External links
- Phosphoinositide 3-kinase C2 family in Pfam
- UMich Orientation of Proteins in Membranes families/superfamily-47 - Orientations of C2 domains in membranes (OPM)
 |

