Biology:Severe acute respiratory syndrome coronavirus
| Severe acute respiratory syndrome coronavirus | |
|---|---|
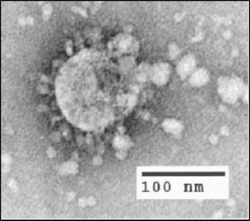
| |
| Electron microscope image of SARS virion | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Genus: | Betacoronavirus |
| Subgenus: | Sarbecovirus |
| Species: | |
| Strain: | Severe acute respiratory syndrome coronavirus
|
| Synonyms | |
| |
Severe Acute respiratory syndrome coronavirus (SARS-CoV or SARS-CoV-1)[2] is a strain of virus that causes severe acute respiratory syndrome (SARS).[3] It is an enveloped, positive-sense, single-stranded RNA virus which infects the epithelial cells within the lungs.[4] The virus enters the host cell by binding to angiotensin-converting enzyme 2.[5] It infects humans, bats, and palm civets.[6][7]
On 16 April 2003, following the outbreak of SARS in Asia and secondary cases elsewhere in the world, the World Health Organization (WHO) issued a press release stating that the coronavirus identified by a number of laboratories was the official cause of SARS. The Centers for Disease Control and Prevention (CDC) in the United States and National Microbiology Laboratory (NML) in Canada identified the SARS-CoV-1 genome in April 2003.[8][9] Scientists at Erasmus University in Rotterdam, the Netherlands, demonstrated that the SARS coronavirus fulfilled Koch's postulates, thereby confirming it as the causative agent. In the experiments, macaques infected with the virus developed the same symptoms as human SARS victims.[10]
A similar virus was discovered in December 2019. This virus, named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is the causative pathogen of the ongoing COVID-19 pandemic.[11]
SARS
Severe acute respiratory syndrome (SARS) is the disease caused by SARS-CoV-1. It causes an often severe illness and is marked initially by systemic symptoms of muscle pain, headache, and fever, followed in 2–14 days by the onset of respiratory symptoms,[12] mainly cough, dyspnea, and pneumonia. Another common finding in SARS patients is a decrease in the number of lymphocytes circulating in the blood.[13]
In the SARS outbreak of 2003, about 9% of patients with confirmed SARS-CoV-1 infection died.[14] The mortality rate was much higher for those over 60 years old, with mortality rates approaching 50% for this subset of patients.[14]
Origin and evolutionary history
On 17 March 2003, WHO established a global network of leading laboratories to collaborate in the identification of the causative agent of SARS. Early on, labs in the network narrowed the search to members of the paramyxovirus and coronavirus families. Early findings shared by the labs pointed to coronaviruses with increasing consistency. On 21 March, scientists from the University of Hong Kong announced the isolation of a new virus that was strongly suspected to be the causative agent of SARS.[15]
Epidemiological evidence suggested a zoonotic origin of the virus: more than 33% of the first detected cases of SARS in Guangdong corresponded to animal or food handlers.[16] Seroprevalence studies reinforced this zoonotic link (a high proportion of asymptomatic animal handlers at markets in Guangdong Province had antibodies against SARS-CoV).[16]
On 12 April 2003, scientists working at the Michael Smith Genome Sciences Centre in Vancouver finished mapping the genetic sequence of a coronavirus believed to be linked to SARS. The team was led by Marco Marra and worked in collaboration with the British Columbia Centre for Disease Control and the National Microbiology Laboratory in Winnipeg, Manitoba, using samples from infected patients in Toronto. The map, hailed by the WHO as an important step forward in fighting SARS, is shared with scientists worldwide via the GSC website (see below). Donald Low of Mount Sinai Hospital in Toronto described the discovery as having been made with "unprecedented speed".[17] The sequence of the SARS coronavirus has since been confirmed by other independent groups.
Molecular epidemiological research demonstrated the virus of 2002–2003 south China outbreak and the virus isolated in the same area in late 2003 and early 2004 outbreaks are different, indicating separate species-crossing events.[18] The phylogeny of the outbreak strains shows that the southwestern provinces including Yunnan, Guizhou and Guangxi compare to the human SARS-CoV-1 better than those of the other provinces, but the viruses' evolution is a product of the host interaction and particularity.[19]
In late May 2003, studies from samples of wild animals sold as food in the local market in Guangdong, China, found a strain of SARS coronavirus could be isolated from masked palm civets (Paguma sp.), but the animals did not always show clinical signs. The preliminary conclusion was the SARS virus crossed the xenographic barrier from palm civet to humans, and more than 10,000 masked palm civets were killed in Guangdong Province. The virus was also later found in raccoon dogs (Nyctereuteus sp.), ferret badgers (Melogale spp.), and domestic cats. In 2005, two studies identified a number of SARS-like coronaviruses in Chinese bats.[20][21] Although the bat SARS virus did not replicate in cell culture, in 2008, American researchers[22] altered the genetic structure of bat SARS virus with the human receptor binding domain both in the bat virus and in the mice which demonstrated how zoonosis might occur in evolution.[23] Phylogenetic analysis of these viruses indicated a high probability that SARS coronavirus originated in bats and spread to humans either directly or through animals held in Chinese markets. The bats did not show any visible signs of disease, but are the likely natural reservoirs of SARS-like coronaviruses. In late 2006, scientists from the Chinese Centre for Disease Control and Prevention of University of Hong Kong and the Guangzhou Centre for Disease Control and Prevention established a genetic link between the SARS coronavirus appearing in civets and humans, confirming claims that the virus had jumped across species.[24]
Bats are likely to be the natural reservoir, that is, the host that harbored the pathogen but that does not show no ill effects and serves as a source of infection. No direct progenitor of SARS-CoV was found in bat populations, but WIV16 was found in a cave in Yunnan province, China between 2013 and 2016, and has a 96% genetically similar virus strain. The hypothesis that SARS-CoV-2 emerged through recombinations of bat SARSr-CoVs in the Yunnan cave of WIV16 or in other yet-to-be-identified bat caves is considered highly likely.[25]
Virology
SARS-CoV-1 follows the replication strategy typical of the coronavirus subfamily. The primary human receptor of the virus is angiotensin-converting enzyme 2 (ACE2) and hemaglutinin (HE),[26] first identified in 2003.[27][28]
Human SARS-CoV-1 appears to have had a complex history of recombination between ancestral coronaviruses that were hosted in several different animal groups.[29][30] In order for recombination to happen at least two SARS-CoV-1 genomes must be present in the same host cell. Recombination may occur during genome replication when the RNA polymerase switches from one template to another (copy choice recombination).[30]
SARS-CoV-1 is one of seven known coronaviruses to infect humans. The other six are:[31]
- Human coronavirus 229E (HCoV-229E)
- Human coronavirus NL63 (HCoV-NL63)
- Human coronavirus OC43 (HCoV-OC43)
- Human coronavirus HKU1 (HCoV-HKU1)
- Middle East respiratory syndrome-related coronavirus (MERS-CoV)
- Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)
Treatments
Currently, research is continuing.
Examples of promising therapies include glycyrrhizin (licorice)[32][33][34][35] and favipiravir.
See also
- Carlo Urbani
- Timeline of the SARS outbreak
- SL-CoV-WIV1
References
Citations
- ↑ "ICTV Taxonomy history: Severe acute respiratory syndrome-related coronavirus" (in en). https://talk.ictvonline.org/taxonomy/p/taxonomy-history?taxnode_id=20181868.
- ↑ Neeltje van Doremalen; Trenton Bushmaker; Dylan H. Morris; Myndi G. Holbrook; Amandine Gamble; Brandi N. Williamson; Azaibi Tamin; Jennifer L. Harcourt et al. (17 March 2020). "Aerosol and Surface Stability of SARS-CoV-2 as Compared with SARS-CoV-1". The New England Journal of Medicine 382 (16): 1564–1567. doi:10.1056/NEJMc2004973. PMID 32182409.
- ↑ Thiel, V., ed (2007). Coronaviruses: Molecular and Cellular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-16-5.
- ↑ "Coronaviruses: An Overview of Their Replication and Pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. Clifton, New Jersey, USA. 2015. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2437-0. "SARS-CoV primarily infects epithelial cells within the lung. The virus is capable of entering macrophages and dendritic cells but only leads to an abortive infection [87,88]."
- ↑ Xing-Yi Ge et al. (2013). "Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor". Nature 503 (7477): 535–538. doi:10.1038/nature12711. PMID 24172901. Bibcode: 2013Natur.503..535G.
- ↑ "Global Epidemiology of Bat Coronaviruses". Viruses 11 (2): 174. 20 February 2019. doi:10.3390/v11020174. ISSN 1999-4915. PMID 30791586. "Most notably, horseshoe bats were found to be the reservoir of SARS-like CoVs, while palm civet cats are considered to be the intermediate host for SARS-CoVs [43,44,45].".
- ↑ "Receptor recognition and cross-species infections of SARS coronavirus". Antiviral Research 100 (1): 246–254. October 2013. doi:10.1016/j.antiviral.2013.08.014. ISSN 0166-3542. PMID 23994189. "See Figure 6.".
- ↑ "Remembering SARS: A Deadly Puzzle and the Efforts to Solve It". Centers for Disease Control and Prevention. 11 April 2013. https://www.cdc.gov/about/history/sars/feature.htm.
- ↑ "Coronavirus never before seen in humans is the cause of SARS". United Nations World Health Organization. 16 April 2006. https://www.who.int/mediacentre/releases/2003/pr31/en/.
- ↑ "Aetiology: Koch's postulates fulfilled for SARS virus". Nature 423 (6937): 240. 2003. doi:10.1038/423240a. PMID 12748632. Bibcode: 2003Natur.423..240F.
- ↑ Lau, Susanna K. P.; Luk, Hayes K. H.; Wong, Antonio C. P.; Li, Kenneth S. M.; Zhu, Longchao; He, Zirong; Fung, Joshua; Chan, Tony T. Y. et al. (2020). "Possible Bat Origin of Severe Acute Respiratory Syndrome Coronavirus 2 – Volume 26, Number 7 – July 2020 – Emerging Infectious Diseases journal – CDC" (in en-us). Emerg Infect Dis 26 (7): 1542–1547. doi:10.3201/eid2607.200092. PMID 32315281.
- ↑ "SARS: epidemiology". Respirology (Carlton, Victoria, USA) 8 (Suppl): S9–S14. November 2003. doi:10.1046/j.1440-1843.2003.00518.x. PMID 15018127.
- ↑ "Hematological findings in SARS patients and possible mechanisms". International Journal of Molecular Medicine 14 (2): 311–315. August 2004. doi:10.3892/ijmm.14.2.311. PMID 15254784. http://www.spandidos-publications.com/ijmm/14/2/311.
- ↑ 14.0 14.1 "Severe acute respiratory syndrome (SARS): development of diagnostics and antivirals". Annals of the New York Academy of Sciences 1067 (1): 500–505. May 2006. doi:10.1196/annals.1354.072. PMID 16804033. Bibcode: 2006NYASA1067..500S.
- ↑ "Severe Acute Respiratory Syndrome (SARS) – multi-country outbreak – Update 12". WHO. 27 March 2003. https://www.who.int/csr/don/2003_03_27b/en/.
- ↑ 16.0 16.1 Skowronski, Danuta M.; Astell, Caroline; Brunham, Robert C.; Low, Donald E.; Petric, Martin; Roper, Rachel L.; Talbot, Pierre J.; Tam, Theresa et al. (February 2005). "Severe Acute Respiratory Syndrome (SARS): A Year in Review". Annual Review of Medicine 56 (1): 357–381. doi:10.1146/annurev.med.56.091103.134135. PMID 15660517.
- ↑ "B.C. lab cracks suspected SARS code". Canada: CBC News. April 2003. http://www.cbc.ca/canada/story/2003/04/12/sars_code030412.html.
- ↑ Wang, Lin-Fa et al. “Review of bats and SARS.” Emerging Infectious Diseases vol. 12,12 (2006): 1834–40. National Library of Medicine website doi:10.3201/eid1212.060401
- ↑ Yu, Ping et al. “Geographical structure of bat SARS-related coronaviruses.” Infection, Genetics and Evolution : Journal of Molecular Epidemiology and Evolutionary Genetics in Infectious Diseases vol. 69 (2019): 224–229. National Library of Medicine website doi:10.1016/j.meegid.2019.02.001
- ↑ "Bats are natural reservoirs of SARS-like coronaviruses". Science 310 (5748): 676–679. 2005. doi:10.1126/science.1118391. PMID 16195424. Bibcode: 2005Sci...310..676L. https://zenodo.org/record/3949088.
- ↑ "Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats". Proceedings of the National Academy of Sciences of the United States of America 102 (39): 14040–14045. 2005. doi:10.1073/pnas.0506735102. PMID 16169905. Bibcode: 2005PNAS..10214040L.
- ↑ Becker, Michelle M et al. “Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice.” Proceedings of the National Academy of Sciences of the United States of America vol. 105,50 (2008): 19944-9. doi:10.1073/pnas.0808116105. National Library of Medicine website Retrieved 13 April 2020.
- ↑ National Academies of Sciences, Engineering, and Medicine, Division on Earth and Life Studies, Board on Life Sciences, Board on Chemical Sciences and Technology, Committee on Strategies for Identifying and Addressing Potential Biodefense Vulnerabilities Posed by Synthetic Biology. (5 December 2018). Biodefense in the Age of Synthetic Biology. Washington, DC: National Academies Press. pp. 44–45. ISBN:978-0-309-46518-2 DOI 10.17226/24890. Google Books. Retrieved 13 April 2020.
- ↑ "Scientists prove SARS-civet cat link". China Daily. 23 November 2006. http://www.chinadaily.com.cn/china/2006-11/23/content_740511.htm.
- ↑ Cui, Jie; Li, Fang; Shi, Zheng-Li (March 2019). "Origin and evolution of pathogenic coronaviruses". Nature Reviews Microbiology 17 (3): 181–192. doi:10.1038/s41579-018-0118-9. PMID 30531947.
- ↑ Mesecar, Andrew D.; Ratia, Kiira (2008-06-23). "Viral destruction of cell surface receptors: Fig. 1.". Proceedings of the National Academy of Sciences 105 (26): 8807–8808. doi:10.1073/pnas.0804355105. PMID 18574141.
- ↑ "Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus" (in en). Nature 426 (6965): 450–454. November 2003. doi:10.1038/nature02145. ISSN 0028-0836. PMID 14647384. Bibcode: 2003Natur.426..450L.
- ↑ Bakkers, Mark J.G.; Lang, Yifei; Feitsma, Louris J.; Hulswit, Ruben J.G.; De Poot, Stefanie A.H.; Van Vliet, Arno L.W.; Margine, Irina; De Groot-Mijnes, Jolanda D.F. et al. (2017-03-08). "Betacoronavirus Adaptation to Humans Involved Progressive Loss of Hemagglutinin-Esterase Lectin Activity" (in en). Cell Host & Microbe 21 (3): 356–366. doi:10.1016/j.chom.2017.02.008. ISSN 1931-3128. PMID 28279346.
- ↑ Stanhope MJ, Brown JR, Amrine-Madsen H. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect Genet Evol. 2004 Mar;4(1):15-9. PMID 15019585
- ↑ 30.0 30.1 Zhang XW, Yap YL, Danchin A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch Virol. 2005 Jan;150(1):1–20. Epub 2004 Oct 11. PMID 15480857
- ↑ Cinatl, J.; Morgenstern, B.; Bauer, G.; Chandra, P.; Rabenau, H.; Doerr, H. W. (2003-06-14). "Glycyrrhizin, an active component of liquorice roots, and replication of SARS-associated coronavirus". Lancet 361 (9374): 2045–2046. doi:10.1016/s0140-6736(03)13615-x. ISSN 1474-547X. PMID 12814717. PMC 7112442. https://pubmed.ncbi.nlm.nih.gov/12814717/.
- ↑ Bailly, Christian; Vergoten, Gérard (2020-10-01). "Glycyrrhizin: An alternative drug for the treatment of COVID-19 infection and the associated respiratory syndrome?" (in en). Pharmacology & Therapeutics 214: 107618. doi:10.1016/j.pharmthera.2020.107618. ISSN 0163-7258. PMID 32592716.
- ↑ Pilcher, HelenR. (2003-06-13). "Liquorice may tackle SARS". Nature: news030609–16. doi:10.1038/news030609-16.
- ↑ Hoever, Gerold; Baltina, Lidia; Kondratenko, Rimma; Baltina, Lia; Tolstikov, Genrich A.; Doerr, Hans W.; Cinatl, Jindrich Jr. (2005-01-19). "Antiviral activity of glycyrrhizic acid derivatives against SARS-coronavirus". Journal of Medicinal Chemistry 48 (4): 1256–1259. doi:10.1021/jm0493008. PMID 15715493. https://www.researchgate.net/publication/8018655.
Sources
- "Coronavirus as a possible cause of severe acute respiratory syndrome". The Lancet 361 (9366): 1319–1325. April 2003. doi:10.1016/s0140-6736(03)13077-2. PMID 12711465.
- "Characterization of a Novel Coronavirus Associated with Severe Acute Respiratory Syndrome". Science 300 (5624): 1394–1399. 30 May 2003. doi:10.1126/science.1085952. PMID 12730500. Bibcode: 2003Sci...300.1394R.
- "The Genome Sequence of the SARS-Associated coronavirus". Science 300 (5624): 1399–1404. 30 May 2003. doi:10.1126/science.1085953. PMID 12730501. Bibcode: 2003Sci...300.1399M.
- "Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage". Journal of Molecular Biology 331 (5): 991–1004. 29 August 2003. doi:10.1016/S0022-2836(03)00865-9. PMID 12927536.
- "Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome". Proceedings of the National Academy of Sciences of the United States of America 103 (33): 12546–12551. 15 August 2006. doi:10.1073/pnas.0605438103. PMID 16891412. PMC 1531645. Bibcode: 2006PNAS..10312546Y. https://www.ncbi.nlm.nih.gov/sites/entrez.
- Thiel, V., ed (2007). Coronaviruses: Molecular and Cellular Biology (1st ed.). Caister Academic Press. ISBN 978-1-904455-16-5.
- "Coronavirus Replication and Interaction with Host". Animal Viruses: Molecular Biology. Caister Academic Press. 2008. ISBN 978-1-904455-22-6.
External links
- WHO press release identifying and naming the SARS virus
- The SARS virus genetic map
- Science special on the SARS virus (free content: no registration required)
- U.S. Centers for Disease Control and Prevention (CDC) SARS home
- World Health Organization on alert
Wikidata ☰ Q278567 entry