Biology:TRNA-intron endonuclease
| tRNA-intron lyase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Identifiers | |||||||||
| EC number | 4.6.1.16 | ||||||||
| CAS number | 117444-13-0 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| |||||||||
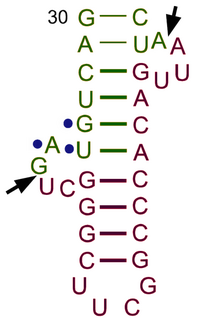
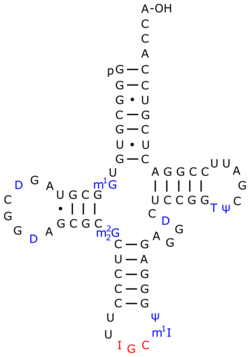
tRNA-intron lyase (EC 4.6.1.16, tRNA intron endonuclease, transfer ribonucleate intron endoribonuclease, tRNA splicing endonuclease, splicing endonuclease, tRNATRPintron endonuclease, transfer splicing endonuclease; systematic name pretRNA lyase (intron-removing; cyclic-2′,3′-phosphate-forming)) is an enzyme.[1][2][3][4] As an endonuclease enzyme, tRNA-intron lyase is responsible for splicing phosphodiester bonds within non-coding ribonucleic acid chains. These non-coding RNA molecules form tRNA molecules after being processed, and this is dependent on tRNA-intron lyase to splice the pretRNA. tRNA processing is an important post-transcriptional modification necessary for tRNA maturation because it locates and removes introns in the pretRNA. This enzyme catalyses the following chemical reaction:
- pretRNA = a 3′-half-tRNA molecule with a 5′-OH end + a 5′-half-tRNA molecule with a 2′,3′-cyclic phosphate end + an intron with a 2′,3′-cyclic phosphate and a 5′-hydroxyl terminus
This enzyme catalyses one of the beginning stages in the maturation of tRNA molecules.
Organisms from every group of the three-domain system relies on tRNA genes. Bacteria does not necessitate cleavage enzymes due to their ability to self-splice. Eukaryotic tRNA-intron endonucleases are hypothesized to evolve from archaeal tRNA-intron endonucleases. Various structures of tRNA-intron lyase have been identified in archaea, less is known about eukaryotic tRNA-intron lyase. The structure of tRNA-intron lyase are maintained by interactions of β strands of local subunits and an electrostatic interaction between a loop and pocket on nearby subunits. Active sites of tRNA-intron lyase are composed of tyrosine, histidine and lysine. Eukaryotes and archaea function similarly and follow the same mechanism to locate unwanted introns and carry out intron splicing. Splicing on both the 3' and 5' ends of the intron.
tRNA-intron lyase requires a level of specificity to the splice site on the pre tRNA. Having a mutation to the splice site on the pre-tRNA could inhibit the enzyme from functioning at the appropriate sites. Although regulating molecules have been defined, current knowledge of understanding of exact mechanisms is limited.
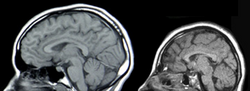
In mammals, including humans, mutations to the tRNA-intron lyase gene are associated with neurodegenerative diseases such as Pontocerebellar Hypoplasias (PCH). This disease leads to atrophy of the cerebellum and pons. This causes microcephaly, severe motor impairment, and severe mental impairment.
Structure
Four types of archaeal tRNA-intron lyase structures have been identified: α4, α′2, (αβ)2, and 𝜀2.[5] All four structures are maintained by two specific interactions.[6] The first interaction is between the β strands of two nearby subunits, and the second interaction is between L10, a negatively charged loop, and the pocket of an adjacent subunit that is positively charged.[6] All four structures feature at least two active sites made of three amino acids (tyrosine, histidine, and lysine) that catalyze the intron splicing reaction.[7]
The homotetrameric tRNA-intron lyase, α4, only found in Methanocaldococcus jannaschii, takes on a rectangular parallelepiped conformation consisting of four α subunits of 179 amino acids each.[6] Each α subunit is made up of five α helices and mixed β sheets. The homodimeric tRNA-intron lyase, α′2 is composed of two α′ subunits that appear to be a fusion protein made up of two α subunits connected by a linker.[8] The α′2 structure is only found in Archaeoglobus fulgidus and Thermoplasma acidophilum.[8] The heterotetramer (αβ)2 structure is found in Nanoarchaeum equitans, Pyrobaculum aerophilum, Aeropyrum pernix, and Methanopyrus kandleri.[9] This structure is composed of two catalytic α subunits and two structural β subunits, with similar function as both of the previously described structures.[9] The last structure, 𝜀2, is a homodimer found only in Candidatus Micrarchaeum acidiphilum.[6] Each subunit is composed of three smaller units: αN, α, and βC joined by two linkers.[6]
Less is known about the eukaryotic tRNA-intron lyase structure. Both yeast (Saccharomyces cerevisiae) and human (Homo sapience, differentiated with “T”) tRNA-intron lyases have four subunits: α (Sen2/TSen2), β (Sen34/TSen34), γ (Sen15/TSen15), and ẟ (Sen54/TSen54).[6] The Sen2 and Sen34 subunits play catalytic roles, while the Sen15 and Sen54 subunits play regulatory roles.[5] Although these structures have been well documented in yeast isoforms using crystallography, only one subunit of the eukaryotic endA enzyme, TSen15, has been elucidated using nuclear magnetic resonance (NMR) spectroscopy.[5]
| Structure Type: | Domain: | Species: | Oligomer: | Subunits: | Subunit Composition: | Image: |
| α4 | Archaea | Methanocaldococcus jannaschii | homotetramer | four α | 5 α helices and mixed β sheets | |
| α′2 | Archaea | Archaeoglobus fulgidus and Thermoplasma acidophilum | homodimer | two α′ | fusion protein with 2 α subunits connected by a linker | |
| (αβ)2 | Archaea | Nanoarchaeum equitans, Pyrobaculum aerophilum, Aeropyrum pernix, and Methanopyrus kandleri | heterotetramer | two α subunits and two β subunits | mixed α helices and β sheets | |
| 𝜀2 | Archaea | Candidatus Micrarchaeum acidiphilum | homodimer | two 𝜀 | Three smaller units (αN, α, and βC) joined by two linkers | |
| αβ𝛾ẟ | Eukarya | Saccharomyces cerevisiae and Homo sapience | heterotetramer | α (Sen2/TSen2), β (Sen34/TSen34), γ (Sen15/TSen15), and ẟ (Sen54/TSen54) | Sen15 linked to Sen34 by β6 pairing |
Function
tRNA-intron endonucleases identify introns along pre-tRNAs and carry out the proper excision mechanism to remove those introns. All three domains of life, Archaea, Bacteria, and Eukarya, contain tRNA genes with introns.[10] Bacterial pre-tRNAs self-splice their introns, whereas Archaea and Eukarya require tRNA-intron endonucleases.[11]
There are two classified groups for Bacterial self-splicing introns, group 1 and group 2. Both groups use an autocatalytic mechanism to remove introns that involves a sequence of transesterification reactions.[12] The only two differences are that group 1 introns use a free external guanosine-5′-triphosphate (GTP) while group 2 doesn't, and the reactions in group 2 introns involve the formation of an intermediate lariat.[12]
The pre-tRNA intron excision mechanism in Eukaryotes and Archaea are shared.[12] There are two different sites at which the tRNA-intron endonuclease cleaves the intron, the 5′- and 3′-splice sites. At the 5’ end, cleavage occurs typically at the canonical position which is one nucleotide 3′ to the anticodon.[13] At the 3’ end, cardinal position 2 (CP2) marks the intron–exon boundary.[11] In Eukarya and Archaea the anticodon-intron (A-I) interaction and bulge-helix-bulge (BHB) motif, respectively, are critical determinants for correct 5′ and 3′ splice-site selection.[11] Moreover, when these two critical determinants are combined, it may be applied to the determination of the correct splice-site selection in archaebacteria.[11] Some introns are found in different parts of tRNA genes, especially in archaeal genomes, but these are minor cases.[13][12] One main difference in function between Eukarya and Archaea is that eukaryal endonucleases typically require the mature domain of the tRNA in order to make accurate cuts in the gene, while archaeal endonucleases do not.[11][12] After cleavage of the two sites, three fragments result: a 3'-half-tRNA molecule with a 5'-OH end, a 5'-half-tRNA molecule with a 2',3'-cyclic phosphate end, and an intron with a 2',3'-cyclic phosphate and a 5'-hydroxyl terminus.
The use of tRNA-intron endonuclease in pre-tRNA intron excision is just one of the steps for tRNA maturation. pre-tRNAs undergo multiple other extensive modifications before the mature tRNA is ready for use in translation.
Evolution
Eukaryotic tRNA-intron endonucleases are considered to have evolved from Archaeal tRNA-intron endonucleases, more specifically the (αβ)2 type.[6] One way this has been shown is through the conservation of the overall structure of Archaeal endonuclease in Eukaryotic endonuclease; The Eukaryotic endonuclease just has two additional subunits (𝛾 and ẟ).[6] Another way this has been shown was done by studying Cyanidioschyzon Merolae, which is a Eukaryote.[6] The eukaryotic organism had tRNA genes with BHB motifs [14] which are only found in archaea and the eukaryal endonucleases are able to recognize and cleave the BHB motifs in vitro.[12]
A convergent evolutionary relationship has also been found between the tRNA-intron endonuclease and RNase A families.[14] RNase A is another enzyme that catalyzes the cleavage of the P–O5' bond of RNA specifically after pyrimidine residues, which is similar to how tRNA-intron endonucleases function and react.[15] The N-terminal domain and C-terminal domain of the endonucleases are similar to the catalytic domains of the LAGLIDADG and PD-(D/E)XK DNases, respectively.[14] This supports the notion of convergent evolution between the endonuclease and RNase A families because none of the other RNase families have been found to be somewhat similar.[14]
Regulation
Specificity
Understanding of the regulation of Eukaryotic tRNA splicing endonuclease is limited, but there are various required conditions for the recognition of splicing cites.[16][17][18] The endonuclease relies on exons in pre-tRNA to determine splice sites.[18] Endonuclease enzymes recognize mature domains in eukaryotic pre-tRNA and determine the distance from the splice site. RNA endonuclease have shown to have conserved base pairing to some introns, despite previous assumptions that introns were not a part of its function.[18] Pairing at these intron sites are necessary for the continuation of the splicing process. Conversely, recognition of splice sites in archaea tRNA only requires the presence of a bulb-helix-bulb motif.[16][4] When the catalytic subunits of the endonuclease recognizes splicing sites in the pre-tRNA, a structural change is prompted to allow splicing.
Regulating Molecules
A recent study has identified a negative regulator of splicing in humans. It was found that the tRNA splicing endonuclease in humans reacts with a polynucleotide kinase known as CLP1.[17] The exact mechanism that CLP1 follows is not entirely understood, but mutations to CLP1 is known to cause neurodegenerative disorders that are similar to those caused by tRNA splicing endonuclease.[17] When CLP1 is not purified by endonuclease, it serves as a negative modulator to the ligation of exons.This inhibits a completed maturation step of tRNA formation that tRNA splicing endonuclease is responsible for. The mechanism would end at intron cleavage. This is regulator is thought to limit or interfere with the continuation of RNA processing.[4]
Pontocerebellar Hypoplasia
Mutations in specific subunits of eukaryotic tRNA-intron lyase are associated with pontocerebellar hypoplasias.[7] Development of this neurodegenerative disease stems from mutations in the TSen54 (ẟ) subunit, TSen2 (α), TSen34 (β), and, more recently, TSen15 (γ).[7] This mutation does not appear in lower-level eukarya, only mammals.[7]
Pontocerebellar hypoplasias (PCH) are a group of neurodegenerative disorders that include atrophy of the cerebellum and pons, microcephaly, severe motor impairment, and severe mental impairment.[19] There are five subtypes of PCH, two of which (PCH2 and PCH4) are linked to mutations in eukaryotic tRNA-intron lyase.[19] PCH2 is associated with symptoms of dystonia and spasticity, while PCH4 is associated with early lethality and a more severe course of disease.[19]
The most common mutation found in patients with PCH2 and PCH4 is a 919G>T mutation in the TSen54 subunit, leading to a substitution at position 307 of alanine by serine.[20] This mutation causes destabilization of the β-β-interaction, altering the ability to form the heterotetramer.[20] The development of PCH from this destabilization is not well understood, but it is thought to be the result of neuronal sensitivity to changes in tRNA levels.[20] Many neuronal diseases result from changes in tRNA and mRNA processing, and this TSen54 subunit mutation may disrupt the tRNA processing pathway.[20]
Other causes of PCH2 and/or PCH4 include other TSen54 point mutations (Q246X, Q343X, S93P), mutations on TSen34 (R58W), and mutations on TSen2 (Y309C).[19] TSen15 was also identified as a potential cause for PCH2, with pathogenic mutations affecting Trp76, His116, and Tyr152.[21]
References
- ↑ "Structural alterations in mutant precursors of the yeast tRNALeu3 gene which behave as defective substrates for a highly purified splicing endoribonuclease". The EMBO Journal 4 (12): 3289–3297. December 1985. doi:10.1002/j.1460-2075.1985.tb04079.x. PMID 3937725.
- ↑ "Precise excision of intervening sequences from precursor tRNAs by a membrane-associated yeast endonuclease". Cell 32 (2): 525–536. February 1983. doi:10.1016/0092-8674(83)90472-5. PMID 6186398.
- ↑ "Transfer RNA intron processing in the halophilic archaebacteria". Canadian Journal of Microbiology 35 (1): 36–42. January 1989. doi:10.1139/m89-006. PMID 2470486.
- ↑ 4.0 4.1 4.2 "A tRNA(Trp) intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties". The Journal of Biological Chemistry 263 (34): 17951–17959. December 1988. doi:10.1016/S0021-9258(19)81308-X. PMID 3192521.
- ↑ 5.0 5.1 5.2 "Three-dimensional structure determined for a subunit of human tRNA splicing endonuclease (Sen15) reveals a novel dimeric fold". Journal of Molecular Biology 366 (1): 155–164. February 2007. doi:10.1016/j.jmb.2006.11.024. PMID 17166513.
- ↑ 6.0 6.1 6.2 6.3 6.4 6.5 6.6 6.7 6.8 "Recent Insights Into the Structure, Function, and Evolution of the RNA-Splicing Endonucleases". Frontiers in Genetics 10: 103. 2019. doi:10.3389/fgene.2019.00103. PMID 30809252.
- ↑ 7.0 7.1 7.2 7.3 "tRNA biology charges to the front". Genes & Development 24 (17): 1832–1860. September 2010. doi:10.1101/gad.1956510. PMID 20810645.
- ↑ 8.0 8.1 "RNA recognition and cleavage by a splicing endonuclease". Science 312 (5775): 906–910. May 2006. doi:10.1126/science.1126629. PMID 16690865. Bibcode: 2006Sci...312..906X.
- ↑ 9.0 9.1 "Crystal structure and assembly of the functional Nanoarchaeum equitans tRNA splicing endonuclease". Nucleic Acids Research 37 (17): 5793–5802. September 2009. doi:10.1093/nar/gkp537. PMID 19578064.
- ↑ "Coevolution of tRNA intron motifs and tRNA endonuclease architecture in Archaea". Proceedings of the National Academy of Sciences of the United States of America 102 (43): 15418–15422. October 2005. doi:10.1073/pnas.0506750102. PMID 16221764. Bibcode: 2005PNAS..10215418T.
- ↑ 11.0 11.1 11.2 11.3 11.4 "Cutting, dicing, healing and sealing: the molecular surgery of tRNA". Wiley Interdisciplinary Reviews. RNA 6 (3): 337–349. May 2015. doi:10.1002/wrna.1279. PMID 25755220.
- ↑ 12.0 12.1 12.2 12.3 12.4 12.5 "Evolution of introns in the archaeal world". Proceedings of the National Academy of Sciences of the United States of America 108 (12): 4782–4787. March 2011. doi:10.1073/pnas.1100862108. PMID 21383132. Bibcode: 2011PNAS..108.4782T.
- ↑ 13.0 13.1 "Handling tRNA introns, archaeal way and eukaryotic way". Frontiers in Genetics 5: 213. 2014-07-10. doi:10.3389/fgene.2014.00213. PMID 25071838.
- ↑ 14.0 14.1 14.2 14.3 "Unusual evolutionary history of the tRNA splicing endonuclease EndA: relationship to the LAGLIDADG and PD-(D/E)XK deoxyribonucleases". Protein Science 10 (3): 656–660. March 2001. doi:10.1110/ps.37101. PMID 11344334.
- ↑ "Ribonuclease a: revealing structure-function relationships with semisynthesis". Journal of the American Chemical Society 117 (31): 8057–8060. August 1995. doi:10.1021/ja00136a001. PMID 21732653.
- ↑ 16.0 16.1 "Diversity and roles of (t)RNA ligases". Cellular and Molecular Life Sciences 69 (16): 2657–2670. August 2012. doi:10.1007/s00018-012-0944-2. PMID 22426497.
- ↑ 17.0 17.1 17.2 "Reconstitution of the human tRNA splicing endonuclease complex: insight into the regulation of pre-tRNA cleavage". Nucleic Acids Research 48 (14): 7609–7622. August 2020. doi:10.1093/nar/gkaa438. PMID 32476018.
- ↑ 18.0 18.1 18.2 "tRNA splicing". The Journal of Biological Chemistry 273 (21): 12685–12688. May 1998. doi:10.1074/jbc.273.21.12685. PMID 9582290.
- ↑ 19.0 19.1 19.2 19.3 "tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia". Nature Genetics 40 (9): 1113–1118. September 2008. doi:10.1038/ng.204. PMID 18711368.
- ↑ 20.0 20.1 20.2 20.3 "Assembly defects of human tRNA splicing endonuclease contribute to impaired pre-tRNA processing in pontocerebellar hypoplasia". Nature Communications 12 (1): 5610. September 2021. doi:10.1038/s41467-021-25870-3. PMID 34584079. Bibcode: 2021NatCo..12.5610S.
- ↑ "Autosomal-Recessive Mutations in the tRNA Splicing Endonuclease Subunit TSEN15 Cause Pontocerebellar Hypoplasia and Progressive Microcephaly". American Journal of Human Genetics 99 (1): 228–235. July 2016. doi:10.1016/j.ajhg.2016.05.023. PMID 27392077.
External links
- TRNA-intron+endonuclease at the US National Library of Medicine Medical Subject Headings (MeSH)
 |