Biology:DNA polymerase II
| DNA Polymerase II | |||||||
|---|---|---|---|---|---|---|---|
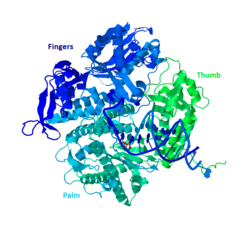
 Crystal structure of DNA pol II (based on PDB entry 3K5M) | |||||||
| Identifiers | |||||||
| Organism | |||||||
| Symbol | polB | ||||||
| Entrez | 944779 | ||||||
| PDB | 3K5M (ECOD) | ||||||
| RefSeq (Prot) | NP_414602.1 | ||||||
| UniProt | P21189 | ||||||
| Other data | |||||||
| EC number | 2.7.7.7 | ||||||
| Chromosome | genome: 0.06 - 0.07 Mb | ||||||
| |||||||
DNA polymerase II (also known as DNA Pol II or Pol II) is a prokaryotic DNA-dependent DNA polymerase encoded by the PolB gene.[1]
DNA Polymerase II is an 89.9-kDa protein and is a member of the B family of DNA polymerases. It was originally isolated by Thomas Kornberg in 1970, and characterized over the next few years.[2][3][4] The in vivo functionality of Pol II is under debate, yet consensus shows that Pol II is primarily involved as a backup enzyme in prokaryotic DNA replication. The enzyme has 5′→3′ DNA synthesis capability as well as 3′→5′ exonuclease proofreading activity. DNA Pol II interacts with multiple binding partners common with DNA Pol III in order to enhance its fidelity and processivity.[1]
Discovery
DNA polymerase I was the first DNA-directed DNA polymerase to be isolated from E. coli.[5] Several studies involving this isolated enzyme indicated that DNA pol I was most likely involved in repair replication and was not the main replicative polymerase.[6] In order to better understand the in vivo role of DNA pol I, E. coli mutants deficient in this enzyme (termed Pol A1−) were generated in 1969 by De Lucia and Cairns.[7] As characterized, this new mutant strain was more sensitive to ultraviolet light, corroborating the hypothesis that DNA pol I was involved in repair replication. The mutant grew at the same rate as the wild type, indicating the presence of another enzyme responsible for DNA replication. The isolation and characterization of this new polymerase involved in semiconservative DNA replication followed, in parallel studies conducted by several labs.[2][3][4] The new polymerase was termed DNA polymerase II, and was believed to be the main replicative enzyme of E. coli for a time.[8] DNA pol II was first crystallized by Anderson et al. in 1994.[9]
In 2023 it was reported that ageing-related accelerated transcription causes Pol II to make more mistakes, leading to flawed copies that can cause numerous diseases.[10]
Structure
DNA Pol II is an 89.9 kD protein, composed of 783 amino acids, that is encoded by the polB (dinA) gene. A globular protein, DNA Pol II functions as a monomer, whereas many other polymerases will form complexes. There are three main sections of this monomer colloquially referred to as the palm, fingers, and thumb. This “hand” closes around a strand of DNA. The palm of the complex contains three catalytic residues that will coordinate with two divalent metal ions in order to function. DNA Pol II has a high quantity of copies in the cell, around 30-50, whereas the level of DNA Pol III in a cell is five times fewer.
Similarity to other group B polymerases
Most of the polymerases have been grouped into families based on similar structure and function. DNA Pol II falls into the Group B along with human DNA Pol α, δ, ϵ, and ζ. These are all homologs of RB69, 9°N-7, and Tgo. The other members of group B do have at least one other subunit which makes the DNA Pol II unique.[11]
Function
Confirmed
Polymerases all are involved with DNA replication in some capacity, synthesizing chains of nucleic acids. DNA replication is a vital aspect of a cell's proliferation. Without replicating its DNA, a cell cannot divide and share its genetic information to progeny. In prokaryotes, like E. coli, DNA Pol III is the major polymerase involved with DNA replication. While DNA Pol II is not a major factor in chromosome replication, it has other roles to fill.
DNA Pol II does participate in DNA replication. While it might not be as fast as DNA Pol III, it has some abilities that make it an effective enzyme. This enzyme has an associated 3′→5′ exonuclease activity along with primase activity. DNA Pol II is a high fidelity enzyme with a substitution error rate of ≤ 2×10−6 and a −1 frameshift error rate of ≤ 1×10−6. DNA Pol II can proofread and process mismatches caused by the Pol III. Banach-Orlowska et al. showed that DNA Pol II is involved with replication but it is strand dependent and preferentially replicates the lagging strand. A proposed mechanism suggests that when DNA Pol III stalls or becomes non-functional, then DNA Pol II is able to be specifically recruited to the replication point and continue replication.[1]
There are many different ways that DNA can be damaged, from UV damage to oxidation, so it is logical that there are different types of polymerases to fix these damages. One important role that DNA Pol II is the major polymerase for the repairing of inter-strand cross-links. Interstrand cross-links are caused by chemicals such as nitrogen mustard and psoralen which create cytotoxic lesions. Repairing these lesions is difficult because both DNA strands have been damaged by the chemical agent and thus the genetic information on both strands is incorrect. The exact mechanism of how these interstrand cross-links are fixed is still being researched, but it is known that Pol II is highly involved.[11]
Activity
DNA Pol II is not the most studied polymerase so there are many proposed functions of this enzyme which are all likely functions but are ultimately unconfirmed:[1]
- repair of DNA damaged by UV irradiation
- replication restart in UV-irradiated E. coli
- adaptive mutagenesis
- long-term survival
Mechanism
During DNA replication, base pairs are subject to damage in the sequence. A damaged sequence of DNA can cause replication to be stalled.[12] In order to fix an error in the sequence, DNA Pol II catalyzes the repair of nucleotide base pairs. In vitro studies have shown that Pol II occasionally interacts with Pol III accessory proteins (β‐clamp and clamp loading complex) giving the Pol II access to the growing nascent strand.[1][13][14][15] Concerning the function of DNA Pol II during DNA replication, this makes sense as any mistakes that Pol III produces will be in the growing strand and not the conservative strand. The N-terminal domain of DNA Pol II is responsible for the association and dissociation of the DNA strand to the catalytic subunit. There are most likely two sites in the N-terminal domain of DNA Pol II that recognize single-stranded DNA. One site(s) is responsible for recruiting single-stranded DNA to DNA Pol II and another site(s) is responsible for the dissociation of single-stranded DNA from DNA Pol II.[16]
Upon binding of substrate, DNA Pol II binds nucleoside triphosphates to maintain the hydrogen bonded structure of DNA. The correct dNTP is then bound and the enzyme complex undergoes conformational changes of subdomains and amino acid residues. These conformational changes allow the rate of repair synthesis to be fast.[17] The active site contains two Mg2+ ions that are stabilized by catalytic Aspartic Acids D419 and D547.[18] Magnesium ions bind to DNA along with dNTP in the open state and coordinate conformational changes of active site amino acid residues in order for catalysis to take place (closed state). After magnesium ions are released, the enzyme returns to its open state.[19]
Species distribution
Prokaryotic
DNA Polymerase II is a member of the polymerase B family and supports Polymerase III in DNA replication moving from the 3′ end to the 5′ end.[20] In the case when Polymerase III stalls during a replication error, Polymerase II can interrupt and excise the mismatched bases. Polymerase II has a much higher fidelity factor than Polymerase III, meaning that it is much less likely to create mispairings. Without Polymerase II's proofreading step, Polymerase III would extend the mispairings and thus create a mutation.[1]
In addition to protecting from mutations that could be caused by Polymerase III, Polymerase II functions to protect against mutations caused by Polymerase IV. Polymerase IV is much more error prone than Polymerase II but also functions to repair mismatched base pairings starting from the 3′ end. Polymerase II protects the 3′ end from Polymerase IV and blocks it from acting. This protection will prevent the formation of mutations while the Polymerase II is functioning normally. If the Polymerase II is knocked out by a mutation or disabled by other factors, Polymerase IV will take its place to fix the mispaired bases.[1]
Eukaryotic
While Polymerase II will not function naturally in conjunction with the eukaryotic members of Family B, it does share similar structural and functional motifs. The members of Family B include Polymerase α, ε, ζ, and δ. These polymerases all function to proofread the newly synthesized DNA in the 3′→5′ direction. These polymerases are capable of synthesizing DNA on both the leading and lagging strands. This class of polymerase tends to be very accurate which allows them to correct any mispairings that occur during DNA synthesis.[20]
Regulation
DNA Polymerase II is naturally abundant in the cell, which usually amounts to five times greater than the amount of Polymerase III. This greater abundance allows Polymerase II to overpower Polymerase III in the case of mispairings. This amount can be increased upon the inducement of the SOS response, which upregulates the polB gene so the amount of Polymerase II increases to about sevenfold greater. Although Polymerase II can work on both strands, it has been shown to prefer the lagging strand versus the leading strand.[1]
See also
References
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 "DNA polymerase II as a fidelity factor in chromosomal DNA synthesis in Escherichia coli". Molecular Microbiology 58 (1): 61–70. October 2005. doi:10.1111/j.1365-2958.2005.04805.x. PMID 16164549.
- ↑ 2.0 2.1 "DNA synthesis in cell-free extracts of a DNA polymerase-defective mutant". Biochemical and Biophysical Research Communications 40 (6): 1348–55. September 1970. doi:10.1016/0006-291X(70)90014-8. PMID 4933688.
- ↑ 3.0 3.1 "A new DNA polymerase activity of Escherichia coli. I. Purification and properties of the activity present in E. coli polA1". Biochemical and Biophysical Research Communications 41 (6): 1557–64. December 1970. doi:10.1016/0006-291X(70)90565-6. PMID 4922636.
- ↑ 4.0 4.1 "DNA polymerase II". Nature 228 (5276): 1050–3. December 1970. doi:10.1038/2281050a0. PMID 4921664. Bibcode: 1970Natur.228.1050K.
- ↑ "Enzymatic synthesis of deoxyribonucleic acid. I. Preparation of substrates and partial purification of an enzyme from Escherichia coli". The Journal of Biological Chemistry 233 (1): 163–70. July 1958. doi:10.1016/S0021-9258(19)68048-8. PMID 13563462.
- ↑ "DNA synthesis in vitro". Nature 226 (5247): 711–3. May 1970. doi:10.1038/226711a0. PMID 4910150. Bibcode: 1970Natur.226..711S.
- ↑ "Isolation of an E. coli strain with a mutation affecting DNA polymerase". Nature 224 (5225): 1164–6. December 1969. doi:10.1038/2241164a0. PMID 4902142. Bibcode: 1969Natur.224.1164D.
- ↑ "Purification and DNA synthesis in cell-free extracts: properties of DNA polymerase II". Proceedings of the National Academy of Sciences of the United States of America 68 (4): 761–4. April 1971. doi:10.1073/pnas.68.4.761. PMID 4927672. Bibcode: 1971PNAS...68..761K.
- ↑ "Crystallization of DNA polymerase II from Escherichia coli". Journal of Molecular Biology 238 (1): 120–2. April 1994. doi:10.1006/jmbi.1994.1765. PMID 8145251.
- ↑ "Ageing-associated changes in transcriptional elongation influence longevity". Nature. 12 April 2023. https://www.nature.com/articles/s41586-023-05922-y. Retrieved 22 May 2023.
- ↑ 11.0 11.1 "Functions of DNA polymerases". Advances in Protein Chemistry 69: 137–65. 2004. doi:10.1016/S0065-3233(04)69005-X. ISBN 9780120342693. PMID 15588842.
- ↑ "Mechanism of DNA polymerase II-mediated frameshift mutagenesis". Proceedings of the National Academy of Sciences of the United States of America 98 (15): 8566–71. July 2001. doi:10.1073/pnas.141113398. PMID 11447256. Bibcode: 2001PNAS...98.8566B.
- ↑ "Conversion of phiX174 viral DNA to double-stranded form by purified Escherichia coli proteins". Proceedings of the National Academy of Sciences of the United States of America 71 (10): 4120–4. October 1974. doi:10.1073/pnas.71.10.4120. PMID 4610569. Bibcode: 1974PNAS...71.4120W.
- ↑ "Escherichia coli DNA polymerase II is stimulated by DNA polymerase III holoenzyme auxiliary subunits". The Journal of Biological Chemistry 266 (7): 4568–73. March 1991. doi:10.1016/S0021-9258(20)64360-5. PMID 1999435. https://www.jbc.org/content/266/7/4568.short.
- ↑ "Processive DNA synthesis by DNA polymerase II mediated by DNA polymerase III accessory proteins". The Journal of Biological Chemistry 267 (16): 11431–8. June 1992. doi:10.1016/S0021-9258(19)49928-6. PMID 1534562. https://www.jbc.org/content/267/16/11431.short.
- ↑ "DNA polymerase II (epsilon) of Saccharomyces cerevisiae dissociates from the DNA template by sensing single-stranded DNA". The Journal of Biological Chemistry 273 (33): 21332–41. August 1998. doi:10.1074/jbc.273.33.21332. PMID 9694894.
- ↑ "Structure and mechanism of DNA polymerase β". Biochemistry 53 (17): 2768–80. May 2014. doi:10.1021/bi500139h. PMID 24717170.
- ↑ "Structural insight into translesion synthesis by DNA Pol II". Cell 139 (7): 1279–89. December 2009. doi:10.1016/j.cell.2009.11.043. PMID 20064374.
- ↑ "Critical role of magnesium ions in DNA polymerase beta's closing and active site assembly". Journal of the American Chemical Society 126 (27): 8441–53. July 2004. doi:10.1021/ja049412o. PMID 15238001.
- ↑ 20.0 20.1 Mandal, Ananya (26 February 2019). "Prokaryotic DNA Polymerases". http://www.news-medical.net/health/Prokaryotic-DNA-Polymerases.aspx.
 |