Biology:Diphosphomevalonate decarboxylase
| diphosphomevalonate decarboxylase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
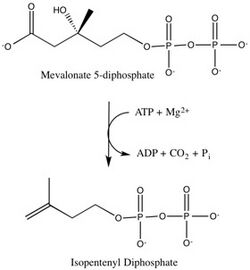 ATP dependent decarboxylation catalyzed by mevalonate diphosphate decarboxylase[1] | |||||||||
| Identifiers | |||||||||
| EC number | 4.1.1.33 | ||||||||
| CAS number | 9024-66-2 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Mevalonate (diphospho) decarboxylase | |
|---|---|
| Identifiers | |
| Symbol | MVD |
| NCBI gene | 4597 |
| HGNC | 7529 |
| OMIM | 603236 |
| RefSeq | NM_002461 |
| UniProt | P53602 |
| Other data | |
| EC number | 4.1.1.33 |
| Locus | Chr. 16 q24.3 |
Diphosphomevalonate decarboxylase (EC 4.1.1.33), most commonly referred to in scientific literature as mevalonate diphosphate decarboxylase[citation needed], is an enzyme that catalyzes the chemical reaction
- ATP + (R)-5-diphosphomevalonate ADP + phosphate + isopentenyl diphosphate + CO2
This enzyme converts mevalonate 5-diphosphate (MVAPP) to isopentenyl diphosphate (IPP) through ATP dependent decarboxylation.[1] The two substrates of this enzyme are ATP and mevalonate 5-diphosphate, whereas its 4 products are ADP, phosphate, isopentenyl diphosphate, and CO2.
Mevalonate diphosphate decarboxylase catalyzes the final step in the mevalonate pathway. The mevalonate pathway is responsible for the biosynthesis of isoprenoids from acetate.[2] This pathway plays a key role in multiple cellular processes by synthesizing sterol isoprenoids, such as cholesterol, and non-sterol isoprenoids, such as dolichol, heme A, tRNA isopentenyltransferase, and ubiquinone.[3][4]
This enzyme belongs to the family of lyases, specifically the carboxy-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is ATP:(R)-5-diphosphomevalonate carboxy-lyase (adding ATP isopentenyl-diphosphate-forming). Other names in common use include pyrophosphomevalonate decarboxylase, mevalonate-5-pyrophosphate decarboxylase, pyrophosphomevalonic acid decarboxylase, 5-pyrophosphomevalonate decarboxylase, mevalonate 5-diphosphate decarboxylase, and ATP:(R)-5-diphosphomevalonate carboxy-lyase (dehydrating).
Enzyme mechanism
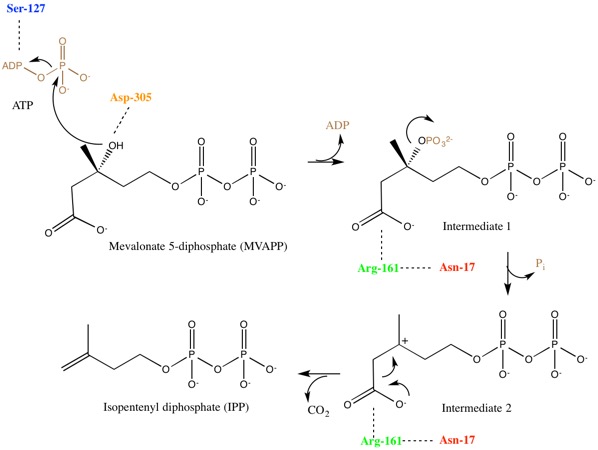
Mevalonate diphosphate decarboxylase recognizes and binds two substrates: ATP and mevalonate 5-diphosphate. After binding, the enzyme performs three types of reactions that can be separated into two main stages. First, phosphorylation occurs. This creates a reactive intermediate, which in the second stage undergoes concerted dephosphorylation and decarboxylation.[5] Many enzyme residues in the active site play important roles in this concerted mechanism. An aspartic acid residue deprotonates the C3 hydroxyl on MVAPP and facilitates the oxygen to attack a phosphate from ATP. As a result, intermediate 1, 3-phosphoMVAPP, now has a much better leaving group, which helps to produce intermediate 2.[1] This third intermediate is a transient beta carboxy carbonium intermediate and provides an "electron sink" that helps drives the decarboxylation reaction.[1]
Enzyme structure

The exact enzyme apparatus of mevalonate diphosphate decarboxylase is not completely understood. Structures of both the yeast and human mevalonate diphosphate decarboxylase have been solved with X-ray crystallography, but scientists have experienced difficulties in obtaining structures of bound metabolites. Scientists have classified mevalonate diphosphate decarboxylase as an enzyme in the GHMP kinase family (galactokinase, homoserine kinase, mevalonate kinase, and phosphomevalonate kinase).[6] Both mevalonate kinase and mevalonate diphosphate decarboxylase probably evolved from a common ancestor since they have a similar fold and catalyze phosphorylation of similar substrates.[6][7] Due to these commonalities, both enzymes are often studied comparatively, and especially in reference to inhibitors.
Though there is limited information, some important residues have been identified and are highlighted in the active site structure and mechanism. Due to the difficulty of obtaining crystal structures of bound substrates, a sulfate ion and water molecules were used to better understand the residues role in substrate binding.[8]
When investigating the human form of mevalonate diphosphate decarboxylase, the following specific residues were identified: arginine-161 (Arg-161), serine-127 (Ser-127), aspartate-305 (Asp-305), and asparagine-17 (Asn-17).[1] Arg-161 interacts with the C1 carbonyl of MVAPP, and Asn-17 is important for hydrogen bonding with this same arginine residue.[1] Asp-305 is positioned about 4 Å from the C3 hydroxyl on MVAPP and acts as a general base catalyst in the active site.[1] Ser-127 aids in orientation of the phosphoryl chain for the phosphate transfer to MVAPP.[1] Mevalonate diphosphate decarboxylase also has a phosphate-binding loop (‘P-loop’) where amino acid residues provide key interactions that stabilize the nucleotide triphosphoryl moiety.[9] The residues from the P-loop are conserved across enzymes in the GHMP kinase family and include Ala-105, Ser-106, Ser-107 and Ala-108.[9]
Biological function
Many different organisms utilize the mevalonate pathway and mevalonate diphosphate decarboxylase, but for different purposes.[9] In gram positive bacteria, isopentenyl diphosphate, the end product of mevalonate diphosphate decarboxylase, is an essential intermediate in peptidoglycan and polyisoprenoid biosynethesis.[9] Therefore, targeting the mevalonate pathway, and mevalonate diphosphate decarboxylase, could be a potential antimicrobial drug.[9]
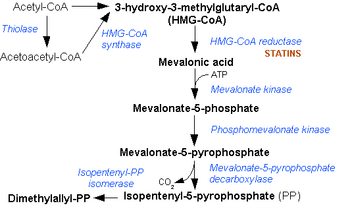
The mevalonate pathway is also used in higher order eukaryotes and plants. Mevalonate diphosphate decarboxylase is mainly present in the liver of mammals where the majority of mevalonate is converted to cholesterol.[10][11] Some of the cholesterol is converted to steroid hormones, bile acids, and vitamin D.[10] Mevalonate is also converted into many other important intermediates in mammalian cells: dolichols (carriers in the assembly of carbohydrate chains in glycoproteins), ubiquinones (important for electron transport), tRNA isopentenyltransferase (used in protein synthesis), and franesylated and geranylgeranylated proteins (membrane associated proteins that appear to be involved in intracellular signaling).[10] The main point of regulation in cholesterol and nonsterol isoprene biosynethsis is HMGCoA reductase, the third enzyme in the mevalonate pathway.[10]
Disease relevance
Coronary artery disease is the leading cause of death in the US general population.[12] Hypercholesterolemia or high cholesterol is considered a major risk factor in coronary artery disease.[13] Therefore, major efforts are focused toward understanding regulation and developing inhibitors of cholesterol biosynthesis.[13] Mevalonate diphosphate decarboxylase is a potential enzyme to be targeted in the cholesterol synthesis pathway. Scientists discovered a molecule, 6-fluoromevalonate (6-FMVA), to be a strong competitive inhibitor of mevalonate diphosphate decarboxylase.[13] The addition of 6-FMVA results in a decrease in cholesterol levels.[13]
Spontaneously hypertensive rats (stroke-prone) (SHRSP) are affected by severe hypertension and cerebral hemorrhage.[14] Scientists have found a low serum cholesterol level in rats with this condition.[14] In SHRSP, mevalonate diphosphate decarboxylase has a much lower activity while HMG-CoA reductase remains unchanged; therefore, mevalonate diphosphate decarboxylase may be responsible for the lower cholesterol biosynthesis in this condition.[14][15] In humans, it is hypothesized that cholesterol deficiency may make the plasma membranes fragile and, as a result, induce angionecrosis in the brain. Reduced serum cholesterol, resulting from a low activity of mevalonate diphosphate decarboxylase, may be the cause of cerebral hemorrhage in some cases.[14]
Structural studies
As of 2015, at least 15 structures have been solved for this class of enzymes, including PDB accession codes 1FI4, 2HK2, 2HK3, and 2HKE.
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 Voynova, NE; Fu, Z; Battaile, KP; Herdendorf, TJ; Kim, JJ; Miziorko, HM (1 December 2008). "Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure.". Archives of Biochemistry and Biophysics 480 (1): 58–67. doi:10.1016/j.abb.2008.08.024. PMID 18823933.
- ↑ Miziorko, HM (15 January 2011). "Enzymes of the mevalonate pathway of isoprenoid biosynthesis.". Archives of Biochemistry and Biophysics 505 (2): 131–43. doi:10.1016/j.abb.2010.09.028. PMID 20932952.
- ↑ Buhaescu, I; Izzedine, H (June 2007). "Mevalonate pathway: a review of clinical and therapeutical implications.". Clinical Biochemistry 40 (9–10): 575–84. doi:10.1016/j.clinbiochem.2007.03.016. PMID 17467679.
- ↑ Miziorko, HM (15 January 2011). "Enzymes of the mevalonate pathway of isoprenoid biosynthesis.". Archives of Biochemistry and Biophysics 505 (2): 131–43. doi:10.1016/j.abb.2010.09.028. PMID 20932952.
- ↑ Byres, E; Alphey, MS; Smith, TK; Hunter, WN (10 August 2007). "Crystal structures of Trypanosoma brucei and Staphylococcus aureus mevalonate diphosphate decarboxylase inform on the determinants of specificity and reactivity.". Journal of Molecular Biology 371 (2): 540–53. doi:10.1016/j.jmb.2007.05.094. PMID 17583736.
- ↑ 6.0 6.1 Qiu, Yongge; Gao, Jinbo; Guo, Fei; Qiao, Yuqin; Li, Ding (November 2007). "Mutation and inhibition studies of mevalonate 5-diphosphate decarboxylase". Bioorganic & Medicinal Chemistry Letters 17 (22): 6164–6168. doi:10.1016/j.bmcl.2007.09.033. PMID 17888661.
- ↑ Qiu, Yongge; Li, Ding (July 2006). "Inhibition of mevalonate 5-diphosphate decarboxylase by fluorinated substrate analogs". Biochimica et Biophysica Acta (BBA) - General Subjects 1760 (7): 1080–1087. doi:10.1016/j.bbagen.2006.03.009. PMID 16626865.
- ↑ Krepkiy, Dmitriy; Miziorko, Henry M. (July 2004). "Identification of active site residues in mevalonate diphosphate decarboxylase: Implications for a family of phosphotransferases". Protein Science 13 (7): 1875–1881. doi:10.1110/ps.04725204. PMID 15169949.
- ↑ 9.0 9.1 9.2 9.3 9.4 Barta, Michael L.; McWhorter, William J.; Miziorko, Henry M.; Geisbrecht, Brian V. (17 July 2012). "Structural Basis for Nucleotide Binding and Reaction Catalysis in Mevalonate Diphosphate Decarboxylase". Biochemistry 51 (28): 5611–5621. doi:10.1021/bi300591x. PMID 22734632.
- ↑ 10.0 10.1 10.2 10.3 Hinson, DD; Chambliss, KL; Toth, MJ; Tanaka, RD; Gibson, KM (November 1997). "Post-translational regulation of mevalonate kinase by intermediates of the cholesterol and nonsterol isoprene biosynthetic pathways.". Journal of Lipid Research 38 (11): 2216–23. doi:10.1016/S0022-2275(20)34935-X. PMID 9392419.
- ↑ Michihara, A; Akasaki, K; Yamori, Y; Tsuji, H (November 2001). "Tissue distribution of a major mevalonate pyrophosphate decarboxylase in rats.". Biological & Pharmaceutical Bulletin 24 (11): 1231–4. doi:10.1248/bpb.24.1231. PMID 11725954.
- ↑ McCullough, P. A. (11 April 2007). "Coronary Artery Disease". Clinical Journal of the American Society of Nephrology 2 (3): 611–616. doi:10.2215/CJN.03871106. PMID 17699471.
- ↑ 14.0 14.1 14.2 14.3 Sawamura, M; Nara, Y; Yamori, Y (25 March 1992). "Liver mevalonate 5-pyrophosphate decarboxylase is responsible for reduced serum cholesterol in stroke-prone spontaneously hypertensive rat.". The Journal of Biological Chemistry 267 (9): 6051–5. doi:10.1016/S0021-9258(18)42660-9. PMID 1556116.
- ↑ Krepkiy, D; Miziorko, HM (July 2004). "Identification of active site residues in mevalonate diphosphate decarboxylase: implications for a family of phosphotransferases.". Protein Science 13 (7): 1875–81. doi:10.1110/ps.04725204. PMID 15169949.
- "Mevalonic acid pyrophosphate and isopentenyl pyrophosphate". J. Biol. Chem. 234 (10): 2595–2604. 1959. doi:10.1016/S0021-9258(18)69744-3. PMID 13801508.
 |

