Biology:Inosine-5′-monophosphate dehydrogenase
| Inosine 5'-monophosphate dehydrogenase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 Structure of IMPDH[1] | |||||||||
| Identifiers | |||||||||
| EC number | 1.1.1.205 | ||||||||
| CAS number | 9028-93-7 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
Inosine-5′-monophosphate dehydrogenase (IMPDH) is a purine biosynthetic enzyme that catalyzes the nicotinamide adenine dinucleotide (NAD+)-dependent oxidation of inosine monophosphate (IMP) to xanthosine monophosphate (XMP), the first committed and rate-limiting step towards the de novo biosynthesis of guanine nucleotides from IMP. IMPDH is a regulator of the intracellular guanine nucleotide pool, and is therefore important for DNA and RNA synthesis, signal transduction, energy transfer, glycoprotein synthesis, as well as other process that are involved in cellular proliferation.
Structure and function
IMPDH is a tetrameric enzyme,[2][3][4] composed of monomeric subunits with molecular masses of approximately 55 kDa[5] and generally consist of 400-500 residues.[6]
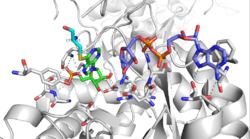
Most IMPDH monomers contain two domains: a catalytic (β/α)8 barrel domain with an active site located in the loops at the C-terminal end of the barrel, and a subdomain consisting of two, repeated cystathionine beta synthetase (CBS) domains that are inserted within the dehydrogenase sequence.[6][8] Monovalent cations have been shown to activate IMPDH enzymes and may serve to stabilize the conformation of the active-site loop.[9]
The CBS domain is not required for catalytic activity. Mutations within the CBS subdomain or a complete deletion of the domains do not impair the in vitro catalytic activity of IMPDH.[10][11] An in vivo deletion of the CBS subdomain in E. coli suggests that the domain can act as a negative transregulator of adenine nucleotide synthesis.[12] IMPDH has also been shown to bind nucleic acids,[13] and this function can be impaired by mutations that are located in the subdomain.[14] The CBS subdomain has also been implicated in mediating IMPDH association with polyribosomes,[15] which suggests a potential moonlighting role for IMPDH as a translational regulatory protein.
Drosophila IMPDH has been demonstrated to act as a sequence-specific transcriptional repressor that can reduce the expression of histone genes and E2F.[16] IMPDH localizes to the nucleus at the end of the S phase and nuclear accumulation is mostly restricted to the G2 phase. In addition, metabolic stress has been shown to induce the nuclear localization of IMPDH.[16]
Mechanism

The overall reaction catalyzed by IMPDH is:[17]
- inosine 5'-phosphate + NAD+ + H2O xanthosine 5'-phosphate + NADH + H+
The mechanism of IMPDH involves a sequence of two different chemical reactions: (1) a fast redox reaction involving a hydride transfer to NAD+ which generates NADH and an enzyme-bound XMP intermediate (E-XMP*) and (2) a hydrolysis step that releases XMP from the enzyme. IMP binds to the active site and a conserved cysteine residue attacks the 2-position of the purine ring. A hydride ion is then transferred from the C2 position to NAD+ and the E-XMP* intermediate is formed. NADH dissociates from the enzyme and a mobile active-site flap element moves a conserved catalytic dyad of arginine and threonine into the newly unoccupied NAD binding site. The arginine residue is thought to act as the general base that activates a water molecule for the hydrolysis reaction.[6] Alternatively, molecular mechanics simulations suggest that in conditions where the arginine residue is protonated, the threonine residue is also capable of activating water by accepting a proton from water while transferring its own proton to a nearby residue.[18]
In Humans
Humans express two distinct isozymes of IMPDH encoded by two distinct genes, IMPDH1 and IMPDH2:
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||
Both isozymes contain 514 residues, have an 84% similarity in peptide sequence, and have similar kinetic properties.[19] Both isozymes are constitutively expressed in most tissues, but IMPDH1 is predominately expressed in the spleen, retina, and peripheral blood leukocytes.[6] IMPDH1 is generally expressed constitutively at low levels, and IMPDH2 is generally upregulated in proliferating cells and neoplastic tissues.[20][21][22] Homozygous IMPDH1 knockout mice demonstrate a mild retinopathy in which a slow, progressive form of retinal degeneration gradually weakens visual transduction,[23] while homozygous IMPDH2 knockout mice display embryonic lethality.[24]
Clinical significance
Guanine nucleotide synthesis is essential for maintaining normal cell function and growth, and is also important for the maintenance of cell proliferation and immune responses. IMPDH expression is found to be upregulated in some tumor tissues and cell lines.[21] B and T lymphocytes display a dependence on IMPDH for normal activation and function,[25][26] and demonstrate upregulated IMPDH expression.[22] Therefore, IMPDH has been addressed as a drug target for immunosuppressive and cancer chemotherapy.
Mycophenolate is an immunosuppressant that is used to prevent transplant rejection and acts through inhibition of IMPDH.
Mutations in the CBS region of IMPDH1 are associated with the RP10 form of autosomal dominant retinitis pigmentosa and dominant Leber's congenital amaurosis.[14]
Research
IMPDH inhibitors have been shown to prevent SARS-CoV-2 replication in cells[27] and are being tested in clinical trials for COVID-19.[28]
See also
References
- ↑ "The immunosuppressive agent mizoribine monophosphate forms a transition state analogue complex with inosine monophosphate dehydrogenase". Biochemistry 42 (4): 857–63. February 2003. doi:10.1021/bi0271401. PMID 12549902.
- ↑ "Characteristics and crystal structure of bacterial inosine-5'-monophosphate dehydrogenase". Biochemistry 38 (15): 4691–700. April 1999. doi:10.1021/bi982858v. PMID 10200156.
- ↑ "Crystal structure of Tritrichomonas foetus inosine-5'-monophosphate dehydrogenase and the enzyme-product complex". Biochemistry 36 (35): 10666–74. September 1997. doi:10.1021/bi9708850. PMID 9271497.
- ↑ "Crystal structures of Tritrichomonasfoetus inosine monophosphate dehydrogenase in complex with substrate, cofactor and analogs: a structural basis for the random-in ordered-out kinetic mechanism". J. Mol. Biol. 326 (2): 517–27. February 2003. doi:10.1016/S0022-2836(02)01383-9. PMID 12559919.
- ↑ "The structure of inosine 5'-monophosphate dehydrogenase and the design of novel inhibitors". Immunopharmacology 47 (2–3): 163–84. May 2000. doi:10.1016/S0162-3109(00)00193-4. PMID 10878288.
- ↑ 6.0 6.1 6.2 6.3 "IMP dehydrogenase: structure, mechanism, and inhibition". Chemical Reviews 109 (7): 2903–28. July 2009. doi:10.1021/cr900021w. PMID 19480389.
- ↑ PDB ID: 1NFB, Risal, D., Strickler M,D., Goldstein, B.M.,The Conformation of NAD Bound to Human Inosine Monophosphate Dehydrogenase Type II.
- ↑ "Enzymes essential for the biosynthesis of nucleic acid guanine; inosine 5'-phosphate dehydrogenase of Aerobacter aerogenes". J. Biol. Chem. 226 (1): 339–50. May 1957. doi:10.1016/S0021-9258(18)64835-5. PMID 13428767.
- ↑ "Monovalent cation activation and kinetic mechanism of inosine 5'-monophosphate dehydrogenase". The Journal of Biological Chemistry 271 (3): 1435–40. January 1996. doi:10.1074/jbc.271.3.1435. PMID 8576135.
- ↑ "Autosomal dominant retinitis pigmentosa mutations in inosine 5'-monophosphate dehydrogenase type I disrupt nucleic acid binding". The Biochemical Journal 390 (Pt 1): 41–7. August 2005. doi:10.1042/BJ20042051. PMID 15882147.
- ↑ "Biochemical analysis of the modular enzyme inosine 5'-monophosphate dehydrogenase". Protein Expression and Purification 17 (2): 282–9. November 1999. doi:10.1006/prep.1999.1136. PMID 10545277.
- ↑ "A regulatory role of the Bateman domain of IMP dehydrogenase in adenylate nucleotide biosynthesis". The Journal of Biological Chemistry 284 (12): 7960–9. March 2009. doi:10.1074/jbc.M808541200. PMID 19153081.
- ↑ "Inosine 5'-monophosphate dehydrogenase binds nucleic acids in vitro and in vivo". The Biochemical Journal 379 (Pt 2): 243–51. April 2004. doi:10.1042/BJ20031585. PMID 14766016.
- ↑ 14.0 14.1 "Spectrum and frequency of mutations in IMPDH1 associated with autosomal dominant retinitis pigmentosa and leber congenital amaurosis". Investigative Ophthalmology & Visual Science 47 (1): 34–42. January 2006. doi:10.1167/iovs.05-0868. PMID 16384941.
- ↑ "IMP dehydrogenase type 1 associates with polyribosomes translating rhodopsin mRNA". The Journal of Biological Chemistry 283 (52): 36354–60. December 2008. doi:10.1074/jbc.M806143200. PMID 18974094.
- ↑ 16.0 16.1 "Metabolic enzyme IMPDH is also a transcription factor regulated by cellular state". Molecular Cell 47 (1): 133–9. July 2012. doi:10.1016/j.molcel.2012.04.030. PMID 22658723.
- ↑ "Cloning and sequence analysis of the human and Chinese hamster inosine-5'-monophosphate dehydrogenase cDNAs". J. Biol. Chem. 263 (30): 15769–72. October 1988. doi:10.1016/S0021-9258(19)37654-9. PMID 2902093.
- ↑ "An enzymatic atavist revealed in dual pathways for water activation". PLOS Biology 6 (8): e206. August 2008. doi:10.1371/journal.pbio.0060206. PMID 18752347.
- ↑ "Two distinct cDNAs for human IMP dehydrogenase". The Journal of Biological Chemistry 265 (9): 5292–5. March 1990. doi:10.1016/S0021-9258(19)34120-1. PMID 1969416.
- ↑ "Tissue-differential expression of two distinct genes for human IMP dehydrogenase (E.C.1.1.1.205)". Life Sciences 54 (24): 1917–26. 1994. doi:10.1016/0024-3205(94)90150-3. PMID 7910933.
- ↑ 21.0 21.1 "Increased inosine-5'-phosphate dehydrogenase gene expression in solid tumor tissues and tumor cell lines". Cancer Research 52 (20): 5826–8. October 1992. doi:10.2172/10148922. PMID 1356621. https://digital.library.unt.edu/ark:/67531/metadc1317672/.
- ↑ 22.0 22.1 Inosine-5'-monophosphate dehydrogenase: regulation of expression and role in cellular proliferation and T lymphocyte activation. Progress in Nucleic Acid Research and Molecular Biology. 61. 1998. pp. 181–209. doi:10.1016/S0079-6603(08)60827-2. ISBN 978-0-12-540061-9.
- ↑ "On the molecular pathology of neurodegeneration in IMPDH1-based retinitis pigmentosa". Human Molecular Genetics 13 (6): 641–50. March 2004. doi:10.1093/hmg/ddh061. PMID 14981049.
- ↑ "Targeted disruption of the inosine 5'-monophosphate dehydrogenase type I gene in mice". Molecular and Cellular Biology 23 (18): 6702–12. September 2003. doi:10.1128/MCB.23.18.6702-6712.2003. PMID 12944494.
- ↑ "Mycophenolic acid inhibits inosine 5'-monophosphate dehydrogenase and suppresses immunoglobulin and cytokine production of B cells". International Immunopharmacology 3 (1): 31–7. January 2003. doi:10.1016/s1567-5769(02)00210-2. PMID 12538032.
- ↑ "Inhibition of T lymphocyte activation in mice heterozygous for loss of the IMPDH II gene". The Journal of Clinical Investigation 106 (4): 599–606. August 2000. doi:10.1172/JCI8669. PMID 10953035.
- ↑ Bojkova, Denisa; Klann, Kevin; Koch, Benjamin; Widera, Marek; Krause, David; Ciesek, Sandra; Cinatl, Jindrich; Münch, Christian (2020-05-14). "Proteomics of SARS-CoV-2-infected host cells reveals therapy targets" (in en). Nature 583 (7816): 469–472. doi:10.1038/s41586-020-2332-7. ISSN 1476-4687. PMID 32408336. Bibcode: 2020Natur.583..469B.
- ↑ Clinical trial number NCT04356677 for "Study to Evaluate the Safety and Efficacy of VIRAZOLE® in Hospitalized Adult Participants With Respiratory Distress Due to COVID-19" at ClinicalTrials.gov
Further reading
- "IMPDH1 gene polymorphisms and association with acute rejection in renal transplant patients". Clin. Pharmacol. Ther. 83 (5): 711–7. May 2008. doi:10.1038/sj.clpt.6100347. PMID 17851563.
 |


