Biology:Severe acute respiratory syndrome-related coronavirus
| Severe acute respiratory syndrome–related coronavirus | |
|---|---|
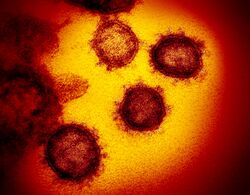
| |
| Transmission electron micrograph of SARS-related coronaviruses emerging from host cells cultured in the lab | |
| Virus classification | |
| Script error: No such module "Taxobox ranks".: | Virus |
| Script error: No such module "Taxobox ranks".: | Riboviria |
| Script error: No such module "Taxobox ranks".: | Orthornavirae |
| Script error: No such module "Taxobox ranks".: | Pisuviricota |
| Script error: No such module "Taxobox ranks".: | Pisoniviricetes |
| Script error: No such module "Taxobox ranks".: | Nidovirales |
| Script error: No such module "Taxobox ranks".: | Coronaviridae |
| Script error: No such module "Taxobox ranks".: | Betacoronavirus |
| Script error: No such module "Taxobox ranks".: | Sarbecovirus |
| Script error: No such module "Taxobox ranks".: | <div style="display:inline" class="script error: no such module "taxobox ranks".">Severe acute respiratory syndrome–related coronavirus |
| Strains | |
| |
| Synonyms | |
| |
Severe acute respiratory syndrome–related coronavirus (SARS-CoV or SARSr-CoV)[note 1] is a species of coronavirus that infects humans, bats and certain other mammals.[2][3] It is an enveloped positive-sense single-stranded RNA virus that enters its host cell by binding to the angiotensin-converting enzyme 2 (ACE2) receptor.[4] It is a member of the genus Betacoronavirus and subgenus Sarbecovirus.[5][6]
Two strains of the virus have caused outbreaks of severe respiratory diseases in humans: severe acute respiratory syndrome coronavirus (SARS-CoV or SARS-CoV-1), which caused the 2002–2004 outbreak of severe acute respiratory syndrome (SARS), and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is causing the 2019–20 pandemic of coronavirus disease 2019 (COVID-19).[7][8] There are hundreds of other strains of SARS-CoV, all of which are only known to infect non-human species: bats are a major reservoir of many strains of SARS-related coronaviruses, and several strains have been identified in palm civets, which were likely ancestors of SARS-CoV.[7][9]
The SARS-related coronavirus was one of several viruses identified by the World Health Organization (WHO) in 2016 as a likely cause of a future epidemic in a new plan developed after the Ebola epidemic for urgent research and development before and during an epidemic towards diagnostic tests, vaccines and medicines. The prediction came to pass with the COVID-19 pandemic.[10][11]
Classification
SARS-related coronavirus is a member of the genus Betacoronavirus (group 2) and subgenus Sarbecovirus (subgroup B).[12] Sarbecoviruses, unlike embecoviruses or alphacoronaviruses, have only one papain-like proteinase (PLpro) instead of two in the open reading frame ORF1.[13] SARSr-CoV was determined to be an early split-off from the betacoronaviruses based on a set of conserved domains that it shares with the group.[14][15]
Bats serve as the main host reservoir species for the SARS-related coronaviruses like SARS-CoV-1 and SARS-CoV-2. The virus has coevolved in the bat host reservoir over a long period of time.[16] Only recently have strains of SARS-related coronavirus evolved and made the cross-species jump from bats to humans, as in the case of the strains SARS-CoV and SARS-CoV-2.[17][4] Both of these strains descended from a single ancestor but made the cross-species jump into humans separately. SARS-CoV-2 is not a direct descendant of SARS-CoV.[7]
Genome
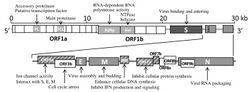
The SARS-related coronavirus is an enveloped, positive-sense, single-stranded RNA virus. Its genome is about 30 kb, which is one of the largest among RNA viruses. The virus has 14 open reading frames which overlap in some cases.[18] The genome has the usual 5′ methylated cap and a 3′ polyadenylated tail.[19] There are 265 nucleotides in the 5'UTR and 342 nucleotides in the 3'UTR.[18]
The 5' methylated cap and 3' polyadenylated tail allows the positive-sense RNA genome to be directly translated by the host cell's ribosome on viral entry.[20] SARSr-CoV is similar to other coronaviruses in that its genome expression starts with translation by the host cell's ribosomes of its initial two large overlapping open reading frames (ORFs), 1a and 1b, both of which produce polyproteins.[18]
| Function of SARS-CoV genome proteins (orf1a to orf9b) | |
|---|---|
| Protein | Function[21][22][23] |
| orf1ab P0C6X7 |
Replicase/transcriptase polyprotein (pp1ab) (nonstructural proteins) |
| orf2 P59594 |
Spike (S) protein, virus binding and entry (structural protein) |
| orf3a P59632 |
Interacts with S, E, M structural proteins; Ion channel activity; Upregulates cytokines and chemokines such as IL-8 and RANTES; Upregulates NF-κB and JNK; Induces apoptosis and cell cycle arrest, via Caspase 8 and -9, and by Bax, p53, and p38 MAP kinase |
| orf3b P59633 |
Upregulates cytokines and chemokines by RUNX1b; Inhibits Type I IFN production and signaling; Induces apoptosis and cell cycle arrest; |
| orf4 P59637 |
Envelope (E) protein, virus assembly and budding (structural protein) |
| orf5 P59596 |
Membrane (M) protein, virus assembly and budding (structural protein) |
| orf6 P59634 |
Enhances cellular DNA synthesis; Inhibits Type I IFN production and signaling |
| orf7a P59635 |
Inhibits cellular protein synthesis; Induces inflammatory response by NF-kappaB and IL-8 promotor; Upregulate chemokines such as IL-8 and RANTES; Upregulates JNK, p38 MAP kinase; Induces apoptosis and cell cycle arrest |
| orf7b Q7TFA1 |
Unknown |
| orf8a Q7TFA0 |
Induces apoptosis through mitochondria pathway |
| orf8b Q80H93 |
Enhances cellular DNA synthesis, also known as X5. |
| orf9a P59595 |
Nucleocapsid (N) protein, viral RNA packaging (structural protein) |
| orf9b P59636 |
Induces apoptosis |
| orf10 Q7TLC7 |
SARS-specific "protein 14" |
The functions of several of the viral proteins are known.[24] ORFs 1a and 1b encode the replicase/transcriptase polyprotein, and later ORFs 2, 4, 5, and 9a encode, respectively, the four major structural proteins: spike, envelope, membrane, and nucleocapsid.[25] The later ORFs also encode for eight unique proteins (orf3a to orf9b), known as the accessory proteins, many with no known homologues. The different functions of the accessory proteins are not well understood.[24]
Morphology
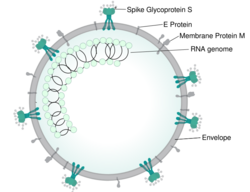
The morphology of the SARS-related coronavirus is characteristic of the coronavirus family as a whole. The viruses are large pleomorphic spherical particles with bulbous surface projections that form a corona around the particles in electron micrographs.[26] The size of the virus particles is in the 80–90 nm range. The envelope of the virus in electron micrographs appears as a distinct pair of electron dense shells.[27]
The viral envelope consists of a lipid bilayer where the membrane (M), envelope (E) and spike (S) proteins are anchored.[28] The spike proteins provide the virus with its bulbous surface projections. The spike protein's interaction with its complement host cell receptor is central in determining the tissue tropism, infectivity, and species range of the virus.[29][30]
Inside the envelope, there is the nucleocapsid, which is formed from multiple copies of the nucleocapsid (N) protein, which are bound to the positive-sense single-stranded (~30 kb) RNA genome in a continuous beads-on-a-string type conformation.[31][32] The lipid bilayer envelope, membrane proteins, and nucleocapsid protect the virus when it is outside the host.[33]
Life cycle
SARS-related coronavirus follows the replication strategy typical of all coronaviruses.[19][34]
Attachment and entry
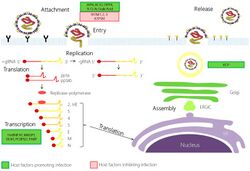
The attachment of the SARS-related coronavirus to the host cell is mediated by the spike protein and its receptor.[35] The spike protein receptor binding domain (RBD) recognizes and attaches to the angiotensin-converting enzyme 2 (ACE2) receptor.[4] Following attachment, the virus can enter the host cell by two different paths. The path the virus takes depends on the host protease available to cleave and activate the receptor-attached spike protein.[36]
The first path the SARS coronavirus can take to enter the host cell is by endocytosis and uptake of the virus in an endosome. The receptor-attached spike protein is then activated by the host's pH-dependent cysteine protease cathepsin L. Activation of the receptor-attached spike protein causes a conformational change, and the subsequent fusion of the viral envelope with the endosomal wall.[36]
Alternatively, the virus can enter the host cell directly by proteolytic cleavage of the receptor-attached spike protein by the host's TMPRSS2 or TMPRSS11D serine proteases at the cell surface.[37][38] In the SARS coronavirus, the activation of the C-terminal part of the spike protein triggers the fusion of the viral envelope with the host cell membrane by inducing conformational changes which are not fully understood.[39]
Genome translation
| Function of coronavirus nonstructural proteins (nsps)[40] | |
|---|---|
| Protein | Function |
| nsp1 | Promotes host mRNA degradation, blocks host translation; blocks innate immune response |
| nsp2 | Binds to prohibitin proteins; unknown function |
| nsp3 | Multidoman transmembrane protein; interacts with N protein; promotes cytokine expression; PLPro domain cleaves polyprotein pp1ab and blocks host's innate immune response; other domains unknown functions |
| nsp4 | Transmembrane scaffold protein; allows proper structure for double membrane vesicles (DMVs) |
| nsp5 | 3CLPro cleaves polyprotein pp1ab |
| nsp6 | Transmembrane scaffold protein; unknown function |
| nsp7 | Forms hexadecameric complex with nsp8; processivity clamp for RdRp (nsp12) |
| nsp8 | Forms hexadecameric complex with nsp7; processivity clamp for RdRp (nsp12); acts as a primase |
| nsp9 | RNA-binding protein (RBP) |
| nsp10 | nsp16 and nsp14 cofactor; forms heterodimer with both; stimulates 2-O-MT (nsp16) and ExoN (nsp14) activity |
| nsp11 | Unknown function |
| nsp12 | RNA-dependent RNA polymerase (RdRp) |
| nsp13 | RNA helicase, 5′ triphosphatase |
| nsp14 | N7 Methyltransferase, 3′-5′ exoribonuclease (ExoN); N7 MTase adds 5′ cap, ExoN proofreads genome |
| nsp15 | Endoribonuclease (NendoU) |
| nsp16 | 2′-O-Methyltransferase (2-O-MT); protects viral RNA from MDA5 |
After fusion the nucleocapsid passes into the cytoplasm, where the viral genome is released.[35] The genome acts as a messenger RNA, and the cell's ribosome translates two-thirds of the genome, which corresponds to the open reading frame ORF1a and ORF1b, into two large overlapping polyproteins, pp1a and pp1ab.
The larger polyprotein pp1ab is a result of a -1 ribosomal frameshift caused by a slippery sequence (UUUAAAC) and a downstream RNA pseudoknot at the end of open reading frame ORF1a.[41] The ribosomal frameshift allows for the continuous translation of ORF1a followed by ORF1b.[42]
The polyproteins contain their own proteases, PLpro and 3CLpro, which cleave the polyproteins at different specific sites. The cleavage of polyprotein pp1ab yields 16 nonstructural proteins (nsp1 to nsp16). Product proteins include various replication proteins such as RNA-dependent RNA polymerase (RdRp), RNA helicase, and exoribonuclease (ExoN).[42]
The two SARS-CoV-2 proteases (PLpro and 3CLpro) also interfere with the immune system response to the viral infection by cleaving three immune system proteins. PLpro cleaves IRF3 and 3CLpro cleaves both NLRP12 and TAB1. "Direct cleavage of IRF3 by NSP3 could explain the blunted Type-I IFN response seen during SARS-CoV-2 infections while NSP5 mediated cleavage of NLRP12 and TAB1 point to a molecular mechanism for enhanced production of IL-6 and inflammatory response observed in COVID-19 patients."[43]
Replication and transcription
File:Replication-transcription complex for Coronaviruses.tif
A number of the nonstructural replication proteins coalesce to form a multi-protein replicase-transcriptase complex (RTC).[42] The main replicase-transcriptase protein is the RNA-dependent RNA polymerase (RdRp). It is directly involved in the replication and transcription of RNA from an RNA strand. The other nonstructural proteins in the complex assist in the replication and transcription process.[40]
The protein nsp14 is a 3'-5' exoribonuclease which provides extra fidelity to the replication process. The exoribonuclease provides a proofreading function to the complex which the RNA-dependent RNA polymerase lacks. Similarly, proteins nsp7 and nsp8 form a hexadecameric sliding clamp as part of the complex which greatly increases the processivity of the RNA-dependent RNA polymerase.[40] The coronaviruses require the increased fidelity and processivity during RNA synthesis because of the relatively large genome size in comparison to other RNA viruses.[44]
One of the main functions of the replicase-transcriptase complex is to transcribe the viral genome. RdRp directly mediates the synthesis of negative-sense subgenomic RNA molecules from the positive-sense genomic RNA. This is followed by the transcription of these negative-sense subgenomic RNA molecules to their corresponding positive-sense mRNAs.[45]
The other important function of the replicase-transcriptase complex is to replicate the viral genome. RdRp directly mediates the synthesis of negative-sense genomic RNA from the positive-sense genomic RNA. This is followed by the replication of positive-sense genomic RNA from the negative-sense genomic RNA.[45]
The replicated positive-sense genomic RNA becomes the genome of the progeny viruses. The various smaller mRNAs are transcripts from the last third of the virus genome which follows the reading frames ORF1a and ORF1b. These mRNAs are translated into the four structural proteins (S, E, M, and N) that will become part of the progeny virus particles and also eight other accessory proteins (orf3 to orf9b) which assist the virus.[46]
Recombination
When two SARS-CoV genomes are present in a host cell, they may interact with each other to form recombinant genomes that can be transmitted to progeny viruses. Recombination likely occurs during genome replication when the RNA polymerase switches from one template to another (copy choice recombination).[47] Human SARS-CoV appears to have had a complex history of recombination between ancestral coronaviruses that were hosted in several different animal groups.[48][47]
Assembly and release
RNA translation occurs inside the endoplasmic reticulum. The viral structural proteins S, E and M move along the secretory pathway into the Golgi intermediate compartment. There, the M proteins direct most protein-protein interactions required for assembly of viruses following its binding to the nucleocapsid.[49]
Progeny viruses are released from the host cell by exocytosis through secretory vesicles.[49]
See also
- SARS-like coronavirus WIV1 (SL-CoV-WIV1)
Notes
- ↑ The terms SARS-CoV and SARSr-CoV are sometimes used interchangeably, especially prior to the discovery of SARS-CoV-2.
References
- ↑ "ICTV Taxonomy history: Severe acute respiratory syndrome-related coronavirus" (in en). https://talk.ictvonline.org/taxonomy/p/taxonomy-history?taxnode_id=20181868.
- ↑ Branswell, Helen (9 November 2015). "SARS-like virus in bats shows potential to infect humans, study finds". Stat News. https://www.statnews.com/2015/11/09/sars-like-virus-bats-shows-potential-infect-humans-study-finds/.
- ↑ "Global Epidemiology of Bat Coronaviruses". Viruses 11 (2): 174. February 2019. doi:10.3390/v11020174. PMID 30791586. "Most notably, horseshoe bats were found to be the reservoir of SARS-like CoVs, while palm civet cats are considered to be the intermediate host for SARS-CoVs [43,44,45].".
- ↑ 4.0 4.1 4.2 "Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor". Nature 503 (7477): 535–8. November 2013. doi:10.1038/nature12711. PMID 24172901. Bibcode: 2013Natur.503..535G.
- ↑ "Virus Taxonomy: 2018 Release" (in en). October 2018. https://talk.ictvonline.org/taxonomy/.
- ↑ "Coronavirus genomics and bioinformatics analysis". Viruses 2 (8): 1804–20. August 2010. doi:10.3390/v2081803. PMID 21994708. "Figure 2. Phylogenetic analysis of RNA-dependent RNA polymerases (Pol) of coronaviruses with complete genome sequences available. The tree was constructed by the neighbor-joining method and rooted using Breda virus polyprotein.".
- ↑ 7.0 7.1 7.2 Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (March 2020). "The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2". Nature Microbiology 5 (4): 536–544. doi:10.1038/s41564-020-0695-z. PMID 32123347.
- ↑ Kohen, Jon; Kupferschmidth, Kai (28 February 2020). "Strategies shift as coronavirus pandemic looms". Science 367 (6481): 962–963. doi:10.1126/science.367.6481.962. PMID 32108093. Bibcode: 2020Sci...367..962C.
- ↑ "Ecoepidemiology and complete genome comparison of different strains of severe acute respiratory syndrome-related Rhinolophus bat coronavirus in China reveal bats as a reservoir for acute, self-limiting infection that allows recombination events". Journal of Virology 84 (6): 2808–19. March 2010. doi:10.1128/JVI.02219-09. PMID 20071579.
- ↑ Kieny, Marie-Paule. "After Ebola, a Blueprint Emerges to Jump-Start R&D". https://blogs.scientificamerican.com/guest-blog/after-ebola-a-blueprint-emerges-to-jump-start-r-d/.
- ↑ "LIST OF PATHOGENS". http://www.who.int/csr/research-and-development/list_of_pathogens/en/.
- ↑ "Global Epidemiology of Bat Coronaviruses". Viruses 11 (2): 174. February 2019. doi:10.3390/v11020174. PMID 30791586. "See Figure 1.".
- ↑ "Coronavirus genomics and bioinformatics analysis". Viruses 2 (8): 1804–20. August 2010. doi:10.3390/v2081803. PMID 21994708. "See Figure 1.".
- ↑ "Coronavirus genomics and bioinformatics analysis". Viruses 2 (8): 1804–20. August 2010. doi:10.3390/v2081803. PMID 21994708. "Furthermore, subsequent phylogenetic analysis using both complete genome sequence and proteomic approaches, it was concluded that SARSr-CoV is probably an early split-off from the Betacoronavirus lineage [1]; See Figure 2.".
- ↑ "Coronaviridae - Figures - Positive Sense RNA Viruses - Positive Sense RNA Viruses (2011)" (in en). International Committee on Taxonomy of Viruses (ICTV). https://talk.ictvonline.org/ictv-reports/ictv_9th_report/positive-sense-rna-viruses-2011/w/posrna_viruses/223/coronaviridae-figures. "See Figure 2."
- ↑ "SARS-Coronavirus ancestor's foot-prints in South-East Asian bat colonies and the refuge theory". Infection, Genetics and Evolution 11 (7): 1690–702. October 2011. doi:10.1016/j.meegid.2011.06.021. PMID 21763784. "Betacoronaviruses-b ancestors, meaning SARSr-CoVs ancestors, could have been historically hosted by the common ancestor of the Rhinolophidae and Hipposideridae and could have later evolved independently in the lineages leading towards Rhinolophidae and Hipposideridae betacoronaviruses.".
- ↑ "Evolutionary relationships between bat coronaviruses and their hosts". Emerging Infectious Diseases 13 (10): 1526–32. October 2007. doi:10.3201/eid1310.070448. PMID 18258002.
- ↑ 18.0 18.1 18.2 "Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage". Journal of Molecular Biology 331 (5): 991–1004. August 2003. doi:10.1016/S0022-2836(03)00865-9. PMID 12927536. "The SARS-CoV genome is ∼29.7 kb long and contains 14 open reading frames (ORFs) flanked by 5′ and 3′-untranslated regions of 265 and 342 nucleotides, respectively (Figure 1).".
- ↑ 19.0 19.1 Maier, Helena Jane; Bickerton, Erica; Britton, Paul, eds (2015). "Coronaviruses: an overview of their replication and pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7.
- ↑ Maier, Helena Jane; Bickerton, Erica; Britton, Paul, eds (2015). An Overview of Their Replication and Pathogenesis; Section 2 Genomic Organization. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7.
- ↑ "The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis". Viruses 4 (11): 2902–23. November 2012. doi:10.3390/v4112902. PMID 23202509. "See Table 1.".
- ↑ "Differential stepwise evolution of SARS coronavirus functional proteins in different host species". BMC Evolutionary Biology 9: 52. March 2009. doi:10.1186/1471-2148-9-52. PMID 19261195.
- ↑ Narayanan, Krishna; Huang, Cheng; Makino, Shinji (April 2008). "SARS coronavirus Accessory Proteins". Virus Research 133 (1): 113–121. doi:10.1016/j.virusres.2007.10.009. ISSN 0168-1702. PMID 18045721. "See Table 1.".
- ↑ 24.0 24.1 "The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis". Viruses 4 (11): 2902–23. November 2012. doi:10.3390/v4112902. PMID 23202509.
- ↑ "Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage". Journal of Molecular Biology 331 (5): 991–1004. August 2003. doi:10.1016/S0022-2836(03)00865-9. PMID 12927536. "See Figure 1.".
- ↑ "Ultrastructural characterization of SARS coronavirus". Emerging Infectious Diseases 10 (2): 320–6. February 2004. doi:10.3201/eid1002.030913. PMID 15030705. "Virions acquired an envelope by budding into the cisternae and formed mostly spherical, sometimes pleomorphic, particles that averaged 78 nm in diameter (Figure 1A).".
- ↑ "Supramolecular architecture of severe acute respiratory syndrome coronavirus revealed by electron cryomicroscopy". Journal of Virology 80 (16): 7918–28. August 2006. doi:10.1128/JVI.00645-06. PMID 16873249. "Particle diameters ranged from 50 to 150 nm, excluding the spikes, with mean particle diameters of 82 to 94 nm; Also See Figure 1 for double shell.".
- ↑ "The molecular biology of coronaviruses". Advances in Virus Research 48: 1–100. 1997. doi:10.1016/S0065-3527(08)60286-9. ISBN 9780120398485. PMID 9233431.
- ↑ The molecular biology of coronaviruses. Advances in Virus Research. 66. Academic Press. 2006-01-01. pp. 193–292. doi:10.1016/S0065-3527(06)66005-3. ISBN 9780120398690. "Nevertheless, the interaction between S protein and receptor remains the principal, if not sole, determinant of coronavirus host species range and tissue tropism."
- ↑ "Origin and evolution of pathogenic coronaviruses". Nature Reviews. Microbiology 17 (3): 181–192. March 2019. doi:10.1038/s41579-018-0118-9. PMID 30531947. "Different SARS-CoV strains isolated from several hosts vary in their binding affinities for human ACE2 and consequently in their infectivity of human cells76,78 (Fig. 6b)".
- ↑ Maier, Helena Jane; Bickerton, Erica; Britton, Paul, eds (2015). An Overview of Their Replication and Pathogenesis; Section 2 Genomic Organization. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. "See section: Virion Structure."
- ↑ "The SARS coronavirus nucleocapsid protein--forms and functions". Antiviral Research 103: 39–50. March 2014. doi:10.1016/j.antiviral.2013.12.009. PMID 24418573. "See Figure 4c.".
- ↑ "A structural analysis of M protein in coronavirus assembly and morphology". Journal of Structural Biology 174 (1): 11–22. April 2011. doi:10.1016/j.jsb.2010.11.021. PMID 21130884. "See Figure 10.".
- ↑ Lal, Sunil K, ed (2010). Molecular Biology of the SARS-Coronavirus. doi:10.1007/978-3-642-03683-5. ISBN 978-3-642-03682-8.
- ↑ 35.0 35.1 Maier, Helena Jane; Bickerton, Erica; Britton, Paul, eds (2015). "Coronaviruses: an overview of their replication and pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. "See section: Coronavirus Life Cycle – Attachment and Entry"
- ↑ 36.0 36.1 "Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research". Antiviral Research 100 (3): 605–14. December 2013. doi:10.1016/j.antiviral.2013.09.028. PMID 24121034. "See Figure 2.".
- ↑ "TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein". Journal of Virology 88 (2): 1293–307. January 2014. doi:10.1128/JVI.02202-13. PMID 24227843. "The SARS-CoV can hijack two cellular proteolytic systems to ensure the adequate processing of its S protein. Cleavage of SARS-S can be facilitated by cathepsin L, a pH-dependent endo-/lysosomal host cell protease, upon uptake of virions into target cell endosomes (25). Alternatively, the type II transmembrane serine proteases (TTSPs) TMPRSS2 and HAT can activate SARS-S, presumably by cleavage of SARS-S at or close to the cell surface, and activation of SARS-S by TMPRSS2 allows for cathepsin L-independent cellular entry (26,–28).".
- ↑ "Coronaviruses - drug discovery and therapeutic options". Nature Reviews. Drug Discovery 15 (5): 327–47. May 2016. doi:10.1038/nrd.2015.37. PMID 26868298. "S is activated and cleaved into the S1 and S2 subunits by other host proteases, such as transmembrane protease serine 2 (TMPRSS2) and TMPRSS11D, which enables cell surface non-endosomal virus entry at the plasma membrane.".
- ↑ "The human coronavirus HCoV-229E S-protein structure and receptor binding". eLife 8. October 2019. doi:10.7554/eLife.51230. PMID 31650956.
- ↑ 40.0 40.1 40.2 Maier, Helena Jane; Bickerton, Erica; Britton, Paul, eds (2015). "Coronaviruses: an overview of their replication and pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. "See Table 2."
- ↑ "The molecular biology of coronaviruses". Advances in Virus Research (Academic Press) 66: 193–292. 2006-01-01. doi:10.1016/S0065-3527(06)66005-3. ISBN 9780120398690. PMID 16877062. "See Figure 8.".
- ↑ 42.0 42.1 42.2 Maier, Helena Jane; Bickerton, Erica; Britton, Paul, eds (2015). "Coronaviruses: an overview of their replication and pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. "See section: Replicase Protein Expression"
- ↑ Mehdi Moustaqil (2020-06-05). SARS-CoV-2 proteases cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species and the search for reservoir hosts. doi:10.1101/2020.06.05.135699. https://www.biorxiv.org/content/10.1101/2020.06.05.135699v1.
- ↑ "Homology-Based Identification of a Mutation in the Coronavirus RNA-Dependent RNA Polymerase That Confers Resistance to Multiple Mutagens". Journal of Virology 90 (16): 7415–28. August 2016. doi:10.1128/JVI.00080-16. PMID 27279608. "Finally, these results, combined with those from previous work (33, 44), suggest that CoVs encode at least three proteins involved in fidelity (nsp12-RdRp, nsp14-ExoN, and nsp10), supporting the assembly of a multiprotein replicase-fidelity complex, as described previously (38).".
- ↑ 45.0 45.1 Maier, Helena Jane; Bickerton, Erica; Britton, Paul, eds (2015). "Coronaviruses: an overview of their replication and pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. "See section: Corona Life Cycle – Replication and Transcription"
- ↑ Maier, Helena Jane; Bickerton, Erica; Britton, Paul, eds (2015). "Coronaviruses: an overview of their replication and pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. "See Figure 1."
- ↑ 47.0 47.1 Zhang XW, Yap YL, Danchin A. Testing the hypothesis of a recombinant origin of the SARS-associated coronavirus. Arch Virol. 2005 Jan;150(1):1-20. Epub 2004 Oct 11. PMID: 15480857
- ↑ Stanhope MJ, Brown JR, Amrine-Madsen H. Evidence from the evolutionary analysis of nucleotide sequences for a recombinant history of SARS-CoV. Infect Genet Evol. 2004 Mar;4(1):15-9. PMID: 15019585
- ↑ 49.0 49.1 Maier, Helena Jane; Bickerton, Erica; Britton, Paul, eds (2015). "Coronaviruses: an overview of their replication and pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. Springer. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. "See section: Coronavirus Life Cycle – Assembly and Release"
Further reading
- "Coronavirus as a possible cause of severe acute respiratory syndrome". Lancet 361 (9366): 1319–25. April 2003. doi:10.1016/s0140-6736(03)13077-2. PMID 12711465.
- "Characterization of a novel coronavirus associated with severe acute respiratory syndrome". Science 300 (5624): 1394–9. May 2003. doi:10.1126/science.1085952. PMID 12730500. Bibcode: 2003Sci...300.1394R.
- "The Genome sequence of the SARS-associated coronavirus". Science 300 (5624): 1399–404. May 2003. doi:10.1126/science.1085953. PMID 12730501. Bibcode: 2003Sci...300.1399M.
- "Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage". Journal of Molecular Biology 331 (5): 991–1004. August 2003. doi:10.1016/S0022-2836(03)00865-9. PMID 12927536.
- "Rewiring the severe acute respiratory syndrome coronavirus (SARS-CoV) transcription circuit: engineering a recombination-resistant genome". Proceedings of the National Academy of Sciences of the United States of America 103 (33): 12546–51. August 2006. doi:10.1073/pnas.0605438103. PMID 16891412. Bibcode: 2006PNAS..10312546Y.
- Coronaviruses: Molecular and Cellular Biology (1st ed.). Caister Academic Press. 2007. ISBN 978-1-904455-16-5.
- "Coronavirus Replication and Interaction with Host". Animal Viruses: Molecular Biology. Caister Academic Press. 2008. ISBN 978-1-904455-22-6.
External links
- WHO press release identifying and naming the SARS virus
- The SARS virus genetic map
- Science special on the SARS virus (free content: no registration required)
- U.S. Centers for Disease Control and Prevention (CDC) SARS home
- World Health Organization on alert
Wikidata ☰ Q278567 entry
