Biology:Marine protists
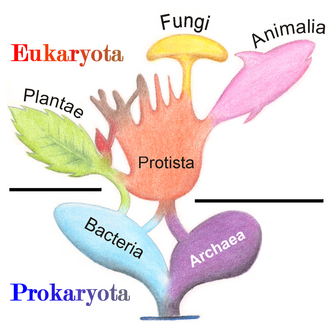
Marine protists are defined by their habitat as protists that live in marine environments, that is, in the saltwater of seas or oceans or the brackish water of coastal estuaries. Life originated as marine single-celled prokaryotes (bacteria and archaea) and later evolved into more complex eukaryotes. Eukaryotes are the more developed life forms known as plants, animals, fungi and protists. Protists are the eukaryotes that cannot be classified as plants, fungi or animals. They are mostly single-celled and microscopic. The term protist came into use historically as a term of convenience for eukaryotes that cannot be strictly classified as plants, animals or fungi. They are not a part of modern cladistics because they are paraphyletic (lacking a common ancestor for all descendants).
Most protists are too small to be seen with the naked eye. They are highly diverse organisms currently organised into 18 phyla, but not easy to classify.[1][2] Studies have shown high protist diversity exists in oceans, deep sea-vents and river sediments, suggesting large numbers of eukaryotic microbial communities have yet to be discovered.[3][4] There has been little research on mixotrophic protists, but recent studies in marine environments found mixotrophic protists contribute a significant part of the protist biomass.[5] Since protists are eukaryotes (and not prokaryotes) they possess within their cell at least one nucleus, as well as organelles such as mitochondria and Golgi bodies. Many protist species can switch between asexual reproduction and sexual reproduction involving meiosis and fertilization.[6]
In contrast to the cells of prokaryotes, the cells of eukaryotes are highly organised. Plants, animals and fungi are usually multi-celled and are typically macroscopic. Most protists are single-celled and microscopic. But there are exceptions. Some single-celled marine protists are macroscopic. Some marine slime molds have unique life cycles that involve switching between unicellular, colonial, and multicellular forms.[7] Other marine protist are neither single-celled nor microscopic, such as seaweed.
Protists have been described as a taxonomic grab bag of misfits where anything that doesn't fit into one of the main biological kingdoms can be placed.[8] Some modern authors prefer to exclude multicellular organisms from the traditional definition of a protist, restricting protists to unicellular organisms.[9][10] This more constrained definition excludes all brown, the multicellular red and green algae, and, sometimes, slime molds (slime molds excluded when multicellularity is defined as "complex").[11]
| Part of a series of overviews on |
| Marine life |
|---|
Background
The ocean represents the largest continuous planetary ecosystem, hosting an enormous variety of organisms, which include microscopic biota such as unicellular eukaryotes (protists). Despite their small size, protists play key roles in marine biogeochemical cycles and harbour tremendous evolutionary diversity.[13][14] Notwithstanding their significance for understanding the evolution of life on Earth and their role in marine food webs, as well as driving biogeochemical cycles to maintain habitability, little is known about their cell biology including reproduction, metabolism and signaling.[12] Most of the biological knowledge available is based on comparison of proteins from cultured species to homologs in genetically tractable model taxa.[15][16][17][18] A main impediment to understanding the cell biology of these diverse eukaryotes is that protocols for genetic modification are available for only a small number of species [19][20] that represent neither the most ecologically relevant protists nor the breadth of eukaryotic diversity. Even so, in the decade to 2020, genome [15][16][17] and transcriptome sequencing initiatives [18] have resulted in nearly 120 million unigenes being identified in protists,[21] which is facilitating the development of genetic tools for model species.[22]

Trophic modes
Protists can be divided broadly into four groups depending on whether their nutrition is plant-like, animal-like, fungal-like,[23] or a mixture of these.[24]
Protists according to how they get food
| |||||||
|---|---|---|---|---|---|---|---|
| Type of protist | Description | Example | Some other examples | ||||
| Plant-like | Algae
(see below) |
Autotrophic protists that make their own food without needing to consume other organisms, usually by photosynthesis (sometimes by chemosynthesis) | 
|
Green algae, Pyramimonas | Red and brown algae, diatoms, coccolithophores and some dinoflagellates. Plant-like protists are important components of phytoplankton discussed below. | ||
| Animal-like | Protozoans
(see below) |
Heterotrophic protists that get their food consuming other organisms (bacteria, archaea and small algae) | 
|
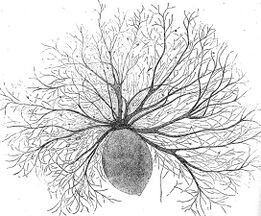
Radiolarian protist as drawn by Haeckel | Foraminiferans, and some marine amoebae, ciliates and flagellates. | ||
| Fungal-like | Slime moulds
and slime nets |
Saprotrophic protists that get their food from the remains of organisms that have broken down and decayed | 
|
Marine slime nets form labyrinthine networks of tubes in which amoeba without pseudopods can travel | Marine lichen | ||
| Mixotrophs | Various
(see below) |
Mixotrophic and osmotrophic protists that get their food from a combination of the above | 
|
Euglena mutabilis, a photosynthetic flagellate | Many marine mixotrops are found among protists, particularly among ciliates and dinoflagellates[5] | ||
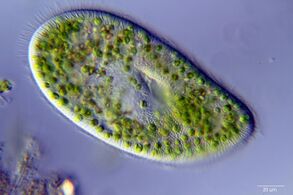
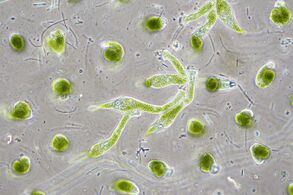
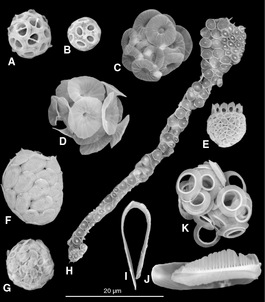
- Single-celled and microscopic protists
Fossil diatom frustule from 32 to 40 mya
Two dinoflagellates
This ciliate is digesting cyanobacteria. The cytostome or mouth is at the bottom right.
The fungus-like protist saprobes are specialized to absorb nutrients from nonliving organic matter, such as dead organisms or their wastes. For instance, many types of oomycetes grow on dead animals or algae. Marine saprobic protists have the essential function of returning inorganic nutrients to the water. This process allows for new algal growth, which in turn generates sustenance for other organisms along the food chain. Indeed, without saprobe species, such as protists, fungi, and bacteria, life would cease to exist as all organic carbon became "tied up" in dead organisms.[27][28]
Mixotrophs
Mixotrophs have no single trophic mode. A mixotroph is an organism that can use a mix of different sources of energy and carbon, instead of having a single trophic mode on the continuum from complete autotrophy at one end to heterotrophy at the other. It is estimated that mixotrophs comprise more than half of all microscopic plankton.[29] There are two types of eukaryotic mixotrophs: those with their own chloroplasts, and those with endosymbionts—and others that acquire them through kleptoplasty or by enslaving the entire phototrophic cell.[30]
The distinction between plants and animals often breaks down in very small organisms. Possible combinations are photo- and chemotrophy, litho- and organotrophy, auto- and heterotrophy or other combinations of these. Mixotrophs can be either eukaryotic or prokaryotic.[31] They can take advantage of different environmental conditions.[32]
Recent studies of marine microzooplankton found 30–45% of the ciliate abundance was mixotrophic, and up to 65% of the amoeboid, foram and radiolarian biomass was mixotrophic.[5]
Phaeocystis is an important algal genus found as part of the marine phytoplankton around the world. It has a polymorphic life cycle, ranging from free-living cells to large colonies.[33] It has the ability to form floating colonies, where hundreds of cells are embedded in a gel matrix, which can increase massively in size during blooms.[34] As a result, Phaeocystis is an important contributor to the marine carbon[35] and sulfur cycles.[36] Phaeocystis species are endosymbionts to acantharian radiolarians.[37][38]
Mixotrophic plankton that combine phototrophy and heterotrophy – table based on Stoecker et al., 2017[39]
| |||||||
|---|---|---|---|---|---|---|---|
| General types | Description | Example | Further examples | ||||
| Bacterioplankton | Photoheterotrophic bacterioplankton | 
|
Vibrio cholerae | Roseobacter spp. Erythrobacter spp. Gammaproteobacterial clade OM60 Widespread among bacteria and archaea | |||
| Phytoplankton | Called constitutive mixotrophs by Mitra et al., 2016.[40] Phytoplankton that eat: photosynthetic protists with inherited plastids and the capacity to ingest prey. | 
|
Ochromonas species | Ochromonas spp. Prymnesium parvum Dinoflagellate examples: Fragilidium subglobosum,Heterocapsa triquetra,Karlodinium veneficum,Neoceratium furca,Prorocentrum minimum | |||
| Zooplankton | Called nonconstitutive mixotrophs by Mitra et al., 2016.[40] Zooplankton that are photosynthetic: microzooplankton or metazoan zooplankton that acquire phototrophy through chloroplast retentiona or maintenance of algal endosymbionts. | ||||||
| Generalists | Protists that retain chloroplasts and rarely other organelles from many algal taxa | 
|
Most oligotrich ciliates that retain plastidsa | ||||
| Specialists | 1. Protists that retain chloroplasts and sometimes other organelles from one algal species or very closely related algal species | 
|
Dinophysis acuminata | Dinophysis spp. Mesodinium rubrum | |||
| 2. Protists or zooplankton with algal endosymbionts of only one algal species or very closely related algal species | 
|
Noctiluca scintillans | Metazooplankton with algal endosymbionts Most mixotrophic Rhizaria (Acantharea, Polycystinea, and Foraminifera) Green Noctiluca scintillans | ||||
| aChloroplast (or plastid) retention = sequestration = enslavement. Some plastid-retaining species also retain other organelles and prey cytoplasm. | |||||||
- Mixoplankton
Tintinnid ciliate Favella
Euglena mutabilis, a photosynthetic flagellate
Protist locomotion
Another way of categorising protists is according to their mode of locomotion. Many unicellular protists, particularly protozoans, are motile and can generate movement using flagella, cilia or pseudopods. Cells which use flagella for movement are usually referred to as flagellates, cells which use cilia are usually referred to as ciliates, and cells which use pseudopods are usually referred to as amoeba or amoeboids. Other protists are not motile, and consequently have no movement mechanism.
Protists according to how they move
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Type of protist | Movement mechanism | Description | Example | Other examples | ||||
| Motile | Flagellates | 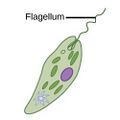
|
A flagellum (Latin for whip) is a lash-like appendage that protrudes from the cell body of some protists (as well as some bacteria). Flagellates use from one to several flagella for locomotion and sometimes as feeding and sensory organelle. | 
|
Cryptophytes | All dinoflagellates and nanoflagellates (choanoflagellates, silicoflagellates, most green algae)[41][42] (Other protists go through a phase as gametes when they have temporary flagellum – some radiolarians, foraminiferans and Apicomplexa)
| ||
| Ciliates | 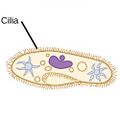
|
A cilium (Latin for eyelash) is a tiny flagellum. Ciliates use multiple cilia, which can number in many hundreds, to power themselves through the water. | 
|
Paramecium bursaria click to see cilia |
Foraminiferans, and some marine amoebae, ciliates and flagellates. | |||
| Amoebas (amoeboids) |
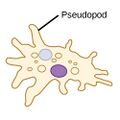
|
Pseudopods (Greek for false feet) are lobe-like appendages which amoebas use to anchor to a solid surface and pull themselves forward. They can change their shape by extending and retracting these pseudopods.[43] | Amoeba | Found in every major protist lineage. Amoeboid cells occur among the protozoans, but also in the algae and the fungi.[44][45] | ||||
| Not motile | none
|
Diatom | Coccolithophores, most diatoms, and non‐motile species of Phaeocystis[42] Among protozoans the parasitic Apicomplexa are non‐motile. | |||||
Flagella are used in prokaryotes (archaea and bacteria) as well as protists. In addition, both flagella and cilia are widely used in eukaryotic cells (plant and animal) apart from protists.
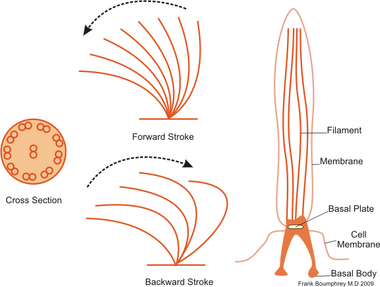
The regular beat patterns of eukaryotic cilia and flagella generates motion on a cellular level. Examples range from the propulsion of single cells such as the swimming of spermatozoa to the transport of fluid along a stationary layer of cells such as in a respiratory tract. Though eukaryotic flagella and motile cilia are ultrastructurally identical, the beating pattern of the two organelles can be different. In the case of flagella, the motion is often planar and wave-like, whereas the motile cilia often perform a more complicated three-dimensional motion with a power and recovery stroke.
Eukaryotic flagella—those of animal, plant, and protist cells—are complex cellular projections that lash back and forth. Eukaryotic flagella are classed along with eukaryotic motile cilia as undulipodia[46] to emphasize their distinctive wavy appendage role in cellular function or motility. Primary cilia are immotile, and are not undulipodia.

Cryptaulax, Abollifer, Bodo, Rhynchomonas, Kiitoksia, Allas, and Metromonas [47]
Ciliates generally have hundreds to thousands of cilia that are densely packed together in arrays. Like the flagella, the cilia are powered by specialised molecular motors. An efficient forward stroke is made with a stiffened flagellum, followed by an inefficient backward stroke made with a relaxed flagellum. During movement, an individual cilium deforms as it uses the high-friction power strokes and the low-friction recovery strokes. Since there are multiple cilia packed together on an individual organism, they display collective behaviour in a metachronal rhythm. This means the deformation of one cilium is in phase with the deformation of its neighbor, causing deformation waves that propagate along the surface of the organism. These propagating waves of cilia are what allow the organism to use the cilia in a coordinated manner to move. A typical example of a ciliated microorganism is the Paramecium, a one-celled, ciliated protozoan covered by thousands of cilia. The cilia beating together allow the Paramecium to propel through the water at speeds of 500 micrometers per second.[48]
- Flagellate, ciliates and amoeba
Green algal flagellate (Chlamydomonas)
Paramecium feeding on bacteria
The ciliate Oxytricha trifallax with cilia clearly visible
Marine algae
Algae is an informal term for a widespread and diverse group of photosynthetic protists which are not necessarily closely related and are thus polyphyletic. Marine algae can be divided into six groups: green, red and brown algae, euglenophytes, dinoflagellates and diatoms.
Dinoflagellates and diatoms are important components of marine algae and have their own sections below. Euglenophytes are a phylum of unicellular flagellates with only a few marine members.
Not all algae are microscopic. Green, red and brown algae all have multicellular macroscopic forms that make up the familiar seaweeds. Green algae, an informal group, contains about 8,000 recognised species.[49] Many species live most of their lives as single cells or are filamentous, while others form colonies made up from long chains of cells, or are highly differentiated macroscopic seaweeds. Red algae, a (disputed) phylum contains about 7,000 recognised species,[50] mostly multicellular and including many notable seaweeds.[50][51] Brown algae form a class containing about 2,000 recognised species,[52] mostly multicellular and including many seaweeds such as kelp. Unlike higher plants, algae lack roots, stems, or leaves. They can be classified by size as microalgae or macroalgae.
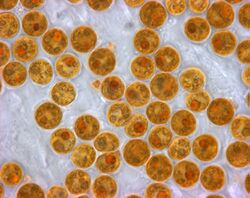
Microalgae are the microscopic types of algae, not visible to the naked eye. They are mostly unicellular species which exist as individuals or in chains or groups, though some are multicellular. Microalgae are important components of the marine protists discussed above, as well as the phytoplankton discussed below. They are very diverse. It has been estimated there are 200,000-800,000 species of which about 50,000 species have been described.[53] Depending on the species, their sizes range from a few micrometers (µm) to a few hundred micrometers. They are specially adapted to an environment dominated by viscous forces.
Chlorella vulgaris, a common green microalgae, in endosymbiosis with a ciliate[54]
Macroalgae are the larger, multicellular and more visible types of algae, commonly called seaweeds. Seaweeds usually grow in shallow coastal waters where they are anchored to the seafloor by a holdfast. Like microalgae, macroalgae (seaweeds) can be regarded as marine protists since they are not true plants. But they are not microorganisms, so they are not within the scope of this article.
Unicellular organisms are usually microscopic, less than one tenth of a millimeter long. There are exceptions. Mermaid's wineglass, a genus of subtropical green algae, is single-celled but remarkably large and complex in form with a single large nucleus, making it a model organism for studying cell biology.[55] Another single-celled algae, Caulerpa taxifolia, has the appearance of a vascular plant including "leaves" arranged neatly up stalks like a fern. Selective breeding in aquariums to produce hardier strains resulted in an accidental release into the Mediterranean where it has become an invasive species known colloquially as killer algae.[56]
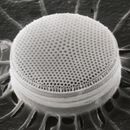
Diatoms
Diatoms are photosynthetic unicellular algae populating the oceans and other waters around the globe. They form a (disputed) phylum containing about 100,000 recognised species. Diatoms generate about 20 per cent of all oxygen produced on the planet each year,[26] and take in over 6.7 billion metric tons of silicon each year from the waters in which they live.[57] They produce 25–45% of the total primary production of organic material in the oceans,[58][59][60] owing to their prevalence in open-ocean regions when total phytoplankton biomass is maximal.[61][62]
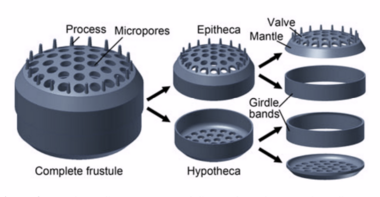
Diatoms are enclosed in protective silica (glass) shells called frustules. They are classified by the shape of these glass cages in which they live, and which they build as they grow. Each frustule is made from two interlocking parts covered with tiny holes through which the diatom exchanges nutrients and wastes.[63] Dead diatoms drift to the ocean floor where, over millions of years, the remains of their frustules can build up as much as half a mile deep.[64] Diatoms have relatively high sinking speeds compared with other phytoplankton groups, and they account for about 40% of particulate carbon exported to ocean depths.[60][65][62]
Diatoms are one of the most common types of phytoplankton
Physically driven seasonal enrichments in surface nutrients favour diatom blooms. Anthropogenic climate change will directly affect these seasonal cycles, changing the timing of blooms and diminishing their biomass, which will reduce primary production and CO2 uptake.[67][62] Remote sensing data suggests there was a global decline of diatoms between 1998 and 2012, particularly in the North Pacific, associated with shallowing of the surface mixed layer and lower nutrient concentrations.[68][62]
Guinardia delicatula, a diatom responsible for diatom blooms in the North Sea [69]
There are over 100,000 species of diatoms accounting for 25–45% of the ocean's primary production
Pennate diatom from an Arctic meltpond, infected with two chytrid-like fungal pathogens. Scale bar = 10 µm.[70]
Coccolithophores
Coccolithophores are minute unicellular photosynthetic protists with two flagella for locomotion. Most of them are protected by calcium carbonate shells covered with ornate circular plates or scales called coccoliths. The term coccolithophore derives from the Greek for a seed carrying stone, referring to their small size and the coccolith stones they carry. Under the right conditions they bloom, like other phytoplankton, and can turn the ocean milky white.[72]

The coccolithophore Emiliania huxleyi
Algae bloom of Emiliania huxleyi off the southern coast of England
Dinoflagellates
Dinoflagellates are usually positioned as part of the algae group, and form a phylum of unicellular flagellates with about 2,000 marine species.[75] The name comes from the Greek "dinos" meaning whirling and the Latin "flagellum" meaning a whip or lash. This refers to the two whip-like attachments (flagella) used for forward movement. Most dinoflagellates are protected with red-brown, cellulose armour. Like other phytoplankton, dinoflagellates are r-strategists which under right conditions can bloom and create red tides. Excavates may be the most basal flagellate lineage.[41]
By trophic orientation dinoflagellates are all over the place. Some dinoflagellates are known to be photosynthetic, but a large fraction of these are in fact mixotrophic, combining photosynthesis with ingestion of prey (phagotrophy).[76] Some species are endosymbionts of marine animals and other protists, and play an important part in the biology of coral reefs. Others predate other protozoa, and a few forms are parasitic. Many dinoflagellates are mixotrophic and could also be classified as phytoplankton.
The toxic dinoflagellate Dinophysis acuta acquire chloroplasts from its prey. "It cannot catch the cryptophytes by itself, and instead relies on ingesting ciliates such as the red Mesodinium rubrum, which sequester their chloroplasts from a specific cryptophyte clade (Geminigera/Plagioselmis/Teleaulax)".[39]
Gyrodinium, one of the few naked dinoflagellates which lack armour
Dinoflagellates often live in symbiosis with other organisms. Many nassellarian radiolarians house dinoflagellate symbionts within their tests.[78] The nassellarian provides ammonium and carbon dioxide for the dinoflagellate, while the dinoflagellate provides the nassellarian with a mucous membrane useful for hunting and protection against harmful invaders.[79] There is evidence from DNA analysis that dinoflagellate symbiosis with radiolarians evolved independently from other dinoflagellate symbioses, such as with foraminifera.[80]
Some dinoflagellates are bioluminescent. At night, ocean water can light up internally and sparkle with blue light because of these dinoflagellates.[81][82] Bioluminescent dinoflagellates possess scintillons, individual cytoplasmic bodies which contain dinoflagellate luciferase, the main enzyme involved in the luminescence. The luminescence, sometimes called the phosphorescence of the sea, occurs as brief (0.1 sec) blue flashes or sparks when individual scintillons are stimulated, usually by mechanical disturbances from, for example, a boat or a swimmer or surf.[83]
Tripos muelleri is recognisable by its U-shaped horns
Karenia brevis produces red tides highly toxic to humans[85]
Noctiluca scintillans, a bioluminescent dinoflagellate[86]
Marine protozoans
Protozoans are protists which feed on organic matter such as other microorganisms or organic tissues and debris.[87][88] Historically, the protozoa were regarded as "one-celled animals", because they often possess animal-like behaviours, such as motility and predation, and lack a cell wall, as found in plants and many algae.[89][90] Although the traditional practice of grouping protozoa with animals is no longer considered valid, the term continues to be used in a loose way to identify single-celled organisms that can move independently and feed by heterotrophy.
Marine protozoans include zooflagellates, foraminiferans, radiolarians and some dinoflagellates.
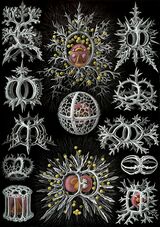
Radiolarians
Radiolarians are unicellular predatory protists encased in elaborate globular shells, typically between 0.1 and 0.2 millimetres in size, usually made of silica and pierced with holes. Their name comes from the Latin for "radius". They catch prey by extending parts of their body through the holes. As with the silica frustules of diatoms, radiolarian shells can sink to the ocean floor when radiolarians die and become preserved as part of the ocean sediment. These remains, as microfossils, provide valuable information about past oceanic conditions.[91]
However acantharian radiolarians have shells made from strontium sulfate crystals
closely replicate some radiolarian shell patterns[92]
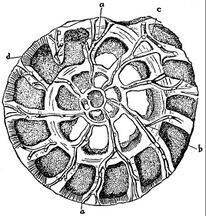
Foraminiferans
Like radiolarians, foraminiferans (forams for short) are single-celled predatory protists, also protected with shells that have holes in them. Their name comes from the Latin for "hole bearers". Their shells, often called tests, are chambered (forams add more chambers as they grow). The shells are usually made of calcite, but are sometimes made of agglutinated sediment particles or chiton, and (rarely) of silica. Most forams are benthic, but about 40 species are planktic.[93] They are widely researched with well established fossil records which allow scientists to infer a lot about past environments and climates.[91]
Live Ammonia tepida streaming granular ectoplasm for catching food
Fossil nummulitid forams of various sizes from the Eocene
The Egyptian pyramids were constructed from limestone that contained nummulites.[94]
A number of forams are mixotrophic (see below). These have unicellular algae as endosymbionts, from diverse lineages such as the green algae, red algae, golden algae, diatoms, and dinoflagellates.[93] Mixotrophic foraminifers are particularly common in nutrient-poor oceanic waters.[95] Some forams are kleptoplastic, retaining chloroplasts from ingested algae to conduct photosynthesis.[96]
Amoeba
Shell or test of a testate amoeba, Arcella sp.
Xenogenic testate amoeba covered in diatoms (from Penard's Amoeba Collection)
File:Amoeba engulfing diatom.ogv
Ciliates
Marine ciliates are major grazers of the phytoplankton.[97][98]
Phytoplankton primary production supports higher trophic levels and fuels microbial remineralization.[99][100] The dominant pelagic grazers of phytoplankton are typically associated with distinct operating modes of the food web compartments and nutrient cycling. Heterotrophic protist grazers and microzooplankton dominance is usually associated with the microbial loop and regenerated production; while mesozooplankton is associated with a linear food chain and export production.[101][102] Grazing on particulate primary production in the global ocean surface is ~10–15% for mesozooplankton and 59–75% for microzooplankton,[103][104][105][106] with estimates for coastal and estuarine systems usually in the a lower range.[106][98]
Ciliates constitute an important component of the microzooplankton community with preference for small-sized preys, in contrast to mesozooplankton, and many ciliate species are also grazed by mesozooplankton.[107] Thus, ciliates can be an important link between small cells and higher trophic levels.[108] Besides their significant role in carbon transfer, ciliates are also considered high quality food, as a source of proteinaceous compounds with a low C:N ratio in comparison to phytoplankton.[109][110][98]
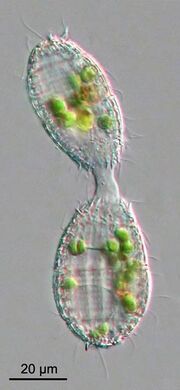
Although many ciliates are heterotrophs, a number of pelagic species are mixotrophic, combining both phagotrophic and phototrophic nutrition (Stoecker, 1998). The recognition of mixotrophy in the marine plankton food web has challenged the classical understanding of pelagic food webs, as autotrophy and heterotrophy are not necessarily two distinct functional compartments.[111] Classical understanding of ecological interactions among plankton, such as competition for nutrients, indicates that nutrient uptake affinity decreases with organism size,[112] favoring smaller sizes under resource limiting conditions. Mixotrophy is advantageous to organisms under nutrient limited conditions, allowing them to reduce direct competition by grazing on smaller prey and increase direct ingestion of nutrients.[113] Modeling results suggest that mixotrophy favors larger organisms, and therefore enhances trophic transfer efficiency.[113][114] On top of that, mixotrophy appears to be important over both, space and time, in marine systems.[115] stressing the need for ecological field studies to further elucidate the role of mixotrophy.[98]
Blepharisma americanum swimming in a drop of pond water with other microorganisms
Macroscopic protists
- Macroscopic protists (see also unicellular macroalgae → )
The single-celled giant amoeba has up to 1000 nuclei and reaches lengths of 5 mm
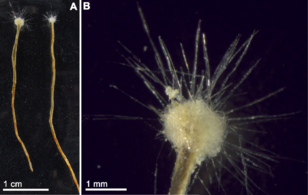
Gromia sphaerica is a large spherical testate amoeba which makes mud trails. Its diameter is up to 3.8 cm.[116]
Spiculosiphon oceana, a unicellular foraminiferan with an appearance and lifestyle that mimics a sponge, grows to 5 cm long.
The xenophyophore, another single-celled foraminiferan, lives in abyssal zones. It has a giant shell up to 20 cm across.[117]
Giant kelp, a brown algae, is not a true plant, yet it is multicellular and can grow to 50m
Planktonic protists
Interactome
Interaction between microbial species has played important roles in evolution and speciation.[118] One of the best examples is that the origin of eukaryotes is grounded in the interaction-events of endosymbiosis; giving rise to mitochondria, chloroplasts, and other metabolic capacities in the eukaryotic cell,[119][120][121][122] Microbial interactions guarantee ecosystem function, having crucial roles in, for instance, carbon channeling in photosymbiosis, control of microalgae blooms by parasites, and phytoplankton-associated bacteria influencing the growth and health of their host.[118]
Despite their importance, understanding of microbial interactions in the ocean and other aquatic systems is rudimentary, and the majority of them are still unknown.[13][123][124][125] The earliest surveys of interactions between aquatic microbes date back to the 19th century. In 1851, while on board HMS Rattlesnake in the Pacific Ocean, Thomas Huxley discovered small yellow–green cells inside the conspicuous planktonic radiolarians which he thought were organelles/[126] Later, Karl Brandt established the yellowish cells were symbiotic alga and named them zooxanthella.[127] Since these early studies, hundreds of others have reported microbial interactions by using classic tools, mainly microscopy, but this knowledge has not yet been gathered into one accessible database. In recent years the high throughput sequencing (HTS)[128][129][130] of environmental DNA or RNA has transformed understanding of microbial diversity [131] and evolution,[132] as well as generating hypotheses on microbial interactions based on correlations of estimated microbial abundances over spatiotemporal scales.[133][134][135][136][118]
The diagram on the right is an overview of the interactions between planktonic protists recorded in a manually curated Protist Interaction DAtabase (PIDA). The network is based on 2422 ecological interactions in the PIDA registered from ~500 publications spanning the last 150 years. The nomenclature and taxonomic order of Eukaryota is based on Adl et al. 2019.[137] The nomenclature and taxonomic order of Bacteria is based on Schultz et al. 2017.[138][118]
The nodes are grouped (outer circle) according to eukaryotic supergroups (or Incertae sedis), Bacteria and Archaea. All major protistan lineages were involved in interactions as hosts, symbionts (mutualists and commensalists), parasites, predators, and/or prey. Predation was the most common interaction (39%), followed by symbiosis (29%), parasitism (18%), and unresolved interactions (14%, where it is uncertain whether the interaction is beneficial or antagonistic). Nodes represent eukaryotic and prokaryotic taxa and are colored accordingly. Node size indicates the number of edges/links that are connected to that node. Each node/taxon is assigned a number, which corresponds with the numbers for taxa in B, C and D. Edges represent interactions between two taxa and are colored according to ecological interaction type: predation (orange), symbiosis (green), and parasitism (purple).[118]
The network is undirected, meaning that a node can contain both parasites/symbionts/prey and hosts/predators. To avoid cluttering of the figure, "Self-loops", which represent cases where both interacting organisms belong to the same taxon (e.g., a dinoflagellate eating another dinoflagellate) are not shown as edges/links in this figure, but are considered in the size of nodes. The outermost circle groups taxa in the different eukaryotic ‘supergroups’ or the prokaryotic domains Bacteria and Archaea. Ancryomonadidae is abbreviated An. Telonema is not placed into any of the supergroups, but classified as Incertae sedis (abbreviated I.S. in the figure). In B, B, and D the following abbreviations for supergroups are used: Ar Archaea, Ba Bacteria, Rh Rhizaria, Al Alveolata, St Stramenopiles, Ha Haptista, Cy Cryptista, Ap Archaeplastida, Ex Excavata, Ob Obazoa, Am Amoebozoa, Cu CRuMS, An Ancryomonadidae, Is Incertae sedis.[118]
B: Predator–prey interactions in PIDA. The node numbers correspond to taxa node numbers in a. Abbreviations for supergroups are described above. Background and nodes are colored according to functional role in the interaction: Prey are colored light orange (left part of figure), while predators are depicted in dark orange (right part of figure). The size of each node represents the number of edges connected to that node.[118]
C. Symbiont–host interactions included in PIDA. The node numbers correspond to node numbers in A. Abbreviations for supergroups are described above. Symbionts are to the left, colored light green, and their hosts are to the right in dark green. The size of each node represents the number of edges connected to that node.[118]
D: Parasite–host interactions included in PIDA. The node numbers correspond to node numbers in A. Abbreviations for supergroups are described above. Parasite taxa are depicted in light purple (left), hosts in dark purple (right).[118]
It was found that protist predators seem to be "multivorous" while parasite–host and symbiont–host interactions appear to have moderate degrees of specialization. The SAR supergroup (i.e., Stramenopiles, Alveolata, and Rhizaria) heavily dominated PIDA, and comparisons against a global-ocean molecular survey (Tara expedition) indicated that several SAR lineages, which are abundant and diverse in the marine realm, were underrepresented among the recorded interactions.[118]
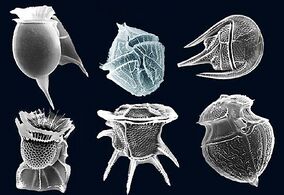
Protist shells
Many protists have protective shells or tests,[139] usually made from calcium carbonate (chalk) or silica (glass). Protists are mostly single-celled and microscopic. Their shells are often tough mineralised forms that resist degradation, and can survive the death of the protist as a microfossil. Although protists are very small, they are ubiquitous. Their numbers are such that their shells play a huge part in the formation of ocean sediments, and in the global cycling of elements and nutrients.
Diatom shells are called frustules and are made from silica. These glass structures have accumulated for over 100 million years leaving rich deposits of nano and microstructured silicon oxide in the form of diatomaceous earth around the globe. The evolutionary causes for the generation of nano and microstructured silica by photosynthetic algae are not yet clear. However, in 2018 it was shown that reflection of ultraviolet light by nanostructured silica protects the DNA in the algal cells, and this may be an evolutionary cause for the formation of the glass cages.[140][141]
Coccolithophores are protected by a shell constructed from ornate circular plates or scales called coccoliths. The coccoliths are made from calcium carbonate or chalk. The term coccolithophore derives from the Greek for a seed carrying stone, referring to their small size and the coccolith stones they carry.[72]
There are benefits for protists that carry protective shells. The diagram on the left above shows some benefits coccolithophore get from carrying coccoliths. In the diagram, (A) represents accelerated photosynthesis including carbon concentrating mechanisms (CCM) and enhanced light uptake via scattering of scarce photons for deep-dwelling species. (B) represents protection from photodamage including sunshade protection from ultraviolet light (UV) and photosynthetic active radiation (PAR) and energy dissipation under high-light conditions. (C) represents armour protection includes protection against viral/bacterial infections and grazing by selective and nonselective grazers.[144]
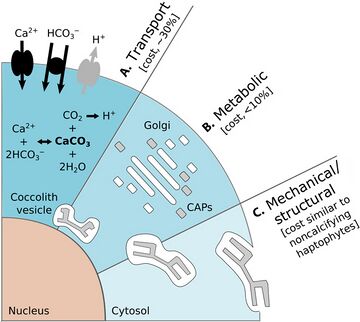
There are also costs for protists that carry protective shells. The diagram on the right above shows some of the energetic costs coccolithophore incur from carrying coccoliths. In the diagram, the energetic costs are reported in percentage of total photosynthetic budget. (A) represents transport processes include the transport into the cell from the surrounding seawater of primary calcification substrates Ca2+ and HCO3− (black arrows) and the removal of the end product H+ from the cell (gray arrow). The transport of Ca2+ through the cytoplasm to the coccolith vesicle (CV) is the dominant cost associated with calcification. (B) represents metabolic processes include the synthesis of coccolith-associated polysaccharides (CAPs – gray rectangles) by the Golgi complex (white rectangles) that regulate the nucleation and geometry of CaCO3 crystals. The completed coccolith (gray plate) is a complex structure of intricately arranged CAPs and CaCO3 crystals. (C) Mechanical and structural processes account for the secretion of the completed coccoliths that are transported from their original position adjacent to the nucleus to the cell periphery, where they are transferred to the surface of the cell.[144]
See also
References
- ↑ Cavalier-Smith T (December 1993). "Kingdom protozoa and its 18 phyla". Microbiological Reviews 57 (4): 953–94. doi:10.1128/mmbr.57.4.953-994.1993. PMID 8302218.
- ↑ Corliss JO (1992). "Should there be a separate code of nomenclature for the protists?". BioSystems 28 (1–3): 1–14. doi:10.1016/0303-2647(92)90003-H. PMID 1292654.
- ↑ "The extent of protist diversity: insights from molecular ecology of freshwater eukaryotes". Proceedings of the Royal Society B: Biological Sciences 272 (1576): 2073–81. 2005. doi:10.1098/rspb.2005.3195. PMID 16191619.
- ↑ "The molecular ecology of microbial eukaryotes unveils a hidden world". Trends in Microbiology 10 (1): 31–8. 2002. doi:10.1016/S0966-842X(01)02257-0. PMID 11755083. http://download.bioon.com.cn/view/upload/month_0803/20080326_daa08a6fdb5d38e3a0d8VBrocN3WtOdR.attach.pdf.
- ↑ 5.0 5.1 5.2 Leles, S.G.; Mitra, A.; Flynn, K.J.; Stoecker, D.K.; Hansen, P.J.; Calbet, A.; McManus, G.B.; Sanders, R.W. et al. (2017). "Oceanic protists with different forms of acquired phototrophy display contrasting biogeographies and abundance". Proceedings of the Royal Society B: Biological Sciences 284 (1860): 20170664. doi:10.1098/rspb.2017.0664. PMID 28768886.
- ↑ Characteristics of Protists In: Rye, Connie; Avissar, Yael; Choi, Jung Ho; DeSaix, Jean; Jurukovski, Vladimir; Wise, Robert R. (2013). Biology.. Houston, Texas. ISBN 978-1-938168-09-3. OCLC 896421272.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Devreotes P (1989). "Dictyostelium discoideum: a model system for cell-cell interactions in development". Science 245 (4922): 1054–8. doi:10.1126/science.2672337. PMID 2672337. Bibcode: 1989Sci...245.1054D.
- ↑ Neil A C, Reece J B, Simon E J (2004) Essential biology with physiology Pearson/Benjamin Cummings, Page 291. ISBN:9780805375039
- ↑ "The other eukaryotes in light of evolutionary protistology". Biology & Philosophy 28 (2): 299–330. 2012. doi:10.1007/s10539-012-9354-y.
- ↑ "The new higher level classification of eukaryotes with emphasis on the taxonomy of protists". The Journal of Eukaryotic Microbiology 52 (5): 399–451. 2005. doi:10.1111/j.1550-7408.2005.00053.x. PMID 16248873.
- ↑ Margulis, Lynn; Chapman, Michael J. (2009-03-19). Kingdoms and Domains: An Illustrated Guide to the Phyla of Life on Earth. Academic Press. ISBN 9780080920146. https://books.google.com/books?id=9IWaqAOGyt4C.
- ↑ 12.0 12.1 Collier, Jackie L.; Rest, Joshua S. (2019). "Swimming, gliding, and rolling toward the mainstream: Cell biology of marine protists". Molecular Biology of the Cell 30 (11): 1245–1248. doi:10.1091/mbc.E18-11-0724. PMID 31084566.
- ↑ 13.0 13.1 Worden, A. Z.; Follows, M. J.; Giovannoni, S. J.; Wilken, S.; Zimmerman, A. E.; Keeling, P. J. (2015). "Rethinking the marine carbon cycle: Factoring in the multifarious lifestyles of microbes". Science 347 (6223). doi:10.1126/science.1257594. PMID 25678667.
- ↑ De Vargas, C. et al. (2015). "Eukaryotic plankton diversity in the sunlit ocean". Science 348 (6237). doi:10.1126/science.1261605. PMID 25999516. https://archimer.ifremer.fr/doc/00270/38135/.
- ↑ 15.0 15.1 Curtis, Bruce A. et al. (2012). "Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs". Nature 492 (7427): 59–65. doi:10.1038/nature11681. PMID 23201678. Bibcode: 2012Natur.492...59C.
- ↑ 16.0 16.1 Armbrust, E. V. et al. (2004). "The Genome of the Diatom Thalassiosira Pseudonana: Ecology, Evolution, and Metabolism". Science 306 (5693): 79–86. doi:10.1126/science.1101156. PMID 15459382. Bibcode: 2004Sci...306...79A. https://digital.library.unt.edu/ark:/67531/metadc885054/.
- ↑ 17.0 17.1 Read, Betsy A. et al. (2013). "Pan genome of the phytoplankton Emiliania underpins its global distribution". Nature 499 (7457): 209–213. doi:10.1038/nature12221. PMID 23760476. Bibcode: 2013Natur.499..209..
- ↑ 18.0 18.1 Keeling, Patrick J. et al. (2014). "The Marine Microbial Eukaryote Transcriptome Sequencing Project (MMETSP): Illuminating the Functional Diversity of Eukaryotic Life in the Oceans through Transcriptome Sequencing". PLOS Biology 12 (6): e1001889. doi:10.1371/journal.pbio.1001889. PMID 24959919.
- ↑ Nymark, Marianne; Sharma, Amit Kumar; Sparstad, Torfinn; Bones, Atle M.; Winge, Per (2016). "A CRISPR/Cas9 system adapted for gene editing in marine algae". Scientific Reports 6: 24951. doi:10.1038/srep24951. PMID 27108533. Bibcode: 2016NatSR...624951N.
- ↑ Hopes, Amanda; Nekrasov, Vladimir; Kamoun, Sophien; Mock, Thomas (2016). "Editing of the urease gene by CRISPR-Cas in the diatom Thalassiosira pseudonana". Plant Methods 12: 49. doi:10.1186/s13007-016-0148-0. PMID 27904648.
- ↑ Carradec, Quentin et al. (2018). "A global ocean atlas of eukaryotic genes". Nature Communications 9 (1): 373. doi:10.1038/s41467-017-02342-1. PMID 29371626. Bibcode: 2018NatCo...9..373C.
- ↑ 22.0 22.1 Faktorová, Drahomíra et al. (2020). "Genetic tool development in marine protists: Emerging model organisms for experimental cell biology". Nature Methods 17 (5): 481–494. doi:10.1038/s41592-020-0796-x. PMID 32251396. 50px Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Whittaker, R.H.; Margulis, L. (1978). "Protist classification and the kingdoms of organisms". Biosystems 10 (1–2): 3–18. doi:10.1016/0303-2647(78)90023-0. PMID 418827.
- ↑ Faure, E; Not, F; Benoiston, AS; Labadie, K; Bittner, L; Ayata, SD (2019). "Mixotrophic protists display contrasted biogeographies in the global ocean". ISME Journal 13 (4): 1072–1083. doi:10.1038/s41396-018-0340-5. PMID 30643201.
- ↑ Budd, Graham E; Jensen, Sören (2017). "The origin of the animals and a 'Savannah' hypothesis for early bilaterian evolution". Biological Reviews 92 (1): 446–473. doi:10.1111/brv.12239. PMID 26588818.
- ↑ 26.0 26.1 The Air You're Breathing? A Diatom Made That
- ↑ Clark M A, Douglas M and Choi J (2018) Biology 2e, 23.4 "Ecology of Protists", OpenStax, Houston, Texas.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Vallet, Marine; Baumeister, Tim U. H.; Kaftan, Filip; Grabe, Veit; Buaya, Anthony; Thines, Marco; Svatoš, Aleš; Pohnert, Georg (2019). "The oomycete Lagenisma coscinodisci hijacks host alkaloid synthesis during infection of a marine diatom". Nature Communications 10 (1): 4938. doi:10.1038/s41467-019-12908-w. PMID 31666506. Bibcode: 2019NatCo..10.4938V.
- ↑ Beware the mixotrophs - they can destroy entire ecosystems 'in a matter of hours'
- ↑ Microscopic body snatchers infest our oceans - Phys.org
- ↑ Eiler A (December 2006). "Evidence for the Ubiquity of Mixotrophic Bacteria in the Upper Ocean: Implications and Consequences". Appl Environ Microbiol 72 (12): 7431–7. doi:10.1128/AEM.01559-06. PMID 17028233. Bibcode: 2006ApEnM..72.7431E.
- ↑ "The mixotroph Ochromonas tuberculata may invade and suppress specialist phago- and phototroph plankton communities depending on nutrient conditions". Oecologia 148 (4): 692–701. July 2006. doi:10.1007/s00442-006-0413-4. PMID 16568278. Bibcode: 2006Oecol.148..692K.
- ↑ Schoemann, Véronique; Becquevort, Sylvie; Stefels, Jacqueline; Rousseau, Véronique; Lancelot, Christiane (2005-01-01). "Phaeocystis blooms in the global ocean and their controlling mechanisms: a review". Journal of Sea Research. Iron Resources and Oceanic Nutrients - Advancement of Global Environmental Simulations 53 (1–2): 43–66. doi:10.1016/j.seares.2004.01.008. Bibcode: 2005JSR....53...43S.
- ↑ "Welcome to the Phaeocystis antarctica genome sequencing project homepage". http://www.phaeocystis.org/.
- ↑ DiTullio, G. R.; Grebmeier, J. M.; Arrigo, K. R.; Lizotte, M. P.; Robinson, D. H.; Leventer, A.; Barry, J. P.; VanWoert, M. L. et al. (2000). "Rapid and early export of Phaeocystis antarctica blooms in the Ross Sea, Antarctica". Nature 404 (6778): 595–598. doi:10.1038/35007061. PMID 10766240. Bibcode: 2000Natur.404..595D.
- ↑ J, Stefels; L, Dijkhuizen; WWC, Gieskes (1995-07-20). "DMSP-lyase activity in a spring phytoplankton bloom off the Dutch coast, related to Phaeocystis sp. abundance". Marine Ecology Progress Series 123: 235–243. doi:10.3354/meps123235. Bibcode: 1995MEPS..123..235S. https://pure.rug.nl/ws/files/62552225/DMSP_lyase_activity_in_a_spring_phytoplankton_bloom_off_the_Dutch_coast.pdf.
- ↑ Decelle, Johan; Simó, Rafel; Galí, Martí; Vargas, Colomban de; Colin, Sébastien; Desdevises, Yves; Bittner, Lucie; Probert, Ian et al. (2012-10-30). "An original mode of symbiosis in open ocean plankton" (in en). Proceedings of the National Academy of Sciences 109 (44): 18000–18005. doi:10.1073/pnas.1212303109. ISSN 0027-8424. PMID 23071304. Bibcode: 2012PNAS..10918000D.
- ↑ Mars Brisbin, Margaret; Grossmann, Mary M.; Mesrop, Lisa Y.; Mitarai, Satoshi (2018). "Intra-host Symbiont Diversity and Extended Symbiont Maintenance in Photosymbiotic Acantharea (Clade F)" (in en). Frontiers in Microbiology 9: 1998. doi:10.3389/fmicb.2018.01998. ISSN 1664-302X. PMID 30210473.
- ↑ 39.0 39.1 Stoecker, D.K.; Hansen, P.J.; Caron, D.A.; Mitra, A. (2017). "Mixotrophy in the marine plankton". Annual Review of Marine Science 9: 311–335. doi:10.1146/annurev-marine-010816-060617. PMID 27483121. Bibcode: 2017ARMS....9..311S.
- ↑ 40.0 40.1 Mitra, AExpression error: Unrecognized word "etal". (2016). "Defining planktonic protist functional groups on mechanisms for energy and nutrient acquisition; incorporation of diverse mixotrophic strategies". Protist 167 (2): 106–20. doi:10.1016/j.protis.2016.01.003. PMID 26927496.
- ↑ 41.0 41.1 Dawson, Scott C; Paredez, Alexander R (2013). "Alternative cytoskeletal landscapes: cytoskeletal novelty and evolution in basal excavate protists". Current Opinion in Cell Biology 25 (1): 134–141. doi:10.1016/j.ceb.2012.11.005. PMID 23312067.
- ↑ 42.0 42.1 Atkinson, A.; Polimene, L.; Fileman, E.S.; Widdicombe, C.E.; McEvoy, A.J.; Smyth, T.J.; Djeghri, N.; Sailley, S.F. et al. (2018). ""Comment. What drives plankton seasonality in a stratifying shelf sea? Some competing and complementary theories""]. Limnology and Oceanography 63 (6): 2877–2884. doi:10.1002/lno.11036. Bibcode: 2018LimOc..63.2877A. http://plymsea.ac.uk/id/eprint/8073/1/Kenitz%20comment_revised%20ACCEPTED.pdf.
- ↑ Singleton, Paul (2006). Dictionary of Microbiology and Molecular Biology, 3rd Edition, revised. Chichester, UK: John Wiley & Sons. pp. 32. ISBN 978-0-470-03545-0. https://archive.org/details/dictionarymicrob00sing_558.
- ↑ David J. Patterson. "Amoebae: Protists Which Move and Feed Using Pseudopodia". Tree of Life web project. http://tolweb.org/notes/?note_id=51.
- ↑ "The Amoebae". The University of Edinburgh. http://www.bms.ed.ac.uk/research/others/smaciver/amoebae.htm.
- ↑ A Dictionary of Biology, 2004, accessed 2011-01-01.
- ↑ Patterson, David J. (2000) "Flagellates: Heterotrophic Protists With Flagella" Tree of Life.
- ↑ Lauga, Eric; Thomas R Powers (25 August 2009). "The hydrodynamics of swimming microorganisms". Reports on Progress in Physics 72 (9): 096601. doi:10.1088/0034-4885/72/9/096601. Bibcode: 2009RPPh...72i6601L.
- ↑ "How many species of algae are there?". Journal of Phycology 48 (5): 1057–63. October 2012. doi:10.1111/j.1529-8817.2012.01222.x. PMID 27011267.
- ↑ 50.0 50.1 Guiry, M.D.; Guiry, G.M. (2016). "Algaebase". http://www.algaebase.org/browse/taxonomy/?id=97240.
- ↑ D. Thomas (2002). Seaweeds. Life Series. Natural History Museum, London. ISBN 978-0-565-09175-0.
- ↑ Hoek, Christiaan; den Hoeck, Hoeck Van; Mann, David; Jahns, H.M. (1995). Algae : an introduction to phycology. Cambridge University Press. p. 166. ISBN 9780521316873. OCLC 443576944. https://books.google.com/books?id=s1P855ZWc0kC&pg=166.
- ↑ Starckx, Senne (31 October 2012) A place in the sun - Algae is the crop of the future, according to researchers in Geel Flanders Today, Retrieved 8 December 2012
- ↑ Duval, B.; Margulis, L. (1995). "The microbial community of Ophrydium versatile colonies: endosymbionts, residents, and tenants". Symbiosis 18: 181–210. PMID 11539474.
- ↑ Mandoli, DF (1998). "Elaboration of Body Plan and Phase Change during Development of Acetabularia: How Is the Complex Architecture of a Giant Unicell Built?". Annual Review of Plant Physiology and Plant Molecular Biology 49: 173–198. doi:10.1146/annurev.arplant.49.1.173. PMID 15012232.
- ↑ Pierre Madl; Maricela Yip (2004). "Literature Review of Caulerpa taxifolia". BUFUS-Info 19 (31). http://biophysics.sbg.ac.at/ct/caulerpa.htm. Retrieved 12 May 2020.
- ↑ Treguer, P.; Nelson, D. M.; Van Bennekom, A. J.; Demaster, D. J.; Leynaert, A.; Queguiner, B. (1995). "The Silica Balance in the World Ocean: A Reestimate". Science 268 (5209): 375–9. doi:10.1126/science.268.5209.375. PMID 17746543. Bibcode: 1995Sci...268..375T.
- ↑ Nelson, David M.; Tréguer, Paul; Brzezinski, Mark A.; Leynaert, Aude; Quéguiner, Bernard (1995). "Production and dissolution of biogenic silica in the ocean: Revised global estimates, comparison with regional data and relationship to biogenic sedimentation". Global Biogeochemical Cycles 9 (3): 359–372. doi:10.1029/95GB01070. Bibcode: 1995GBioC...9..359N.
- ↑ Malviya, Shruti; Scalco, Eleonora; Audic, Stéphane; Vincent, Flora; Veluchamy, Alaguraj; Poulain, Julie; Wincker, Patrick; Iudicone, Daniele et al. (2016). "Insights into global diatom distribution and diversity in the world's ocean". Proceedings of the National Academy of Sciences 113 (11): E1516–E1525. doi:10.1073/pnas.1509523113. PMID 26929361. Bibcode: 2016PNAS..113E1516M.
- ↑ 60.0 60.1 Tréguer, Paul; Bowler, Chris; Moriceau, Brivaela; Dutkiewicz, Stephanie; Gehlen, Marion; Aumont, Olivier; Bittner, Lucie; Dugdale, Richard et al. (2018). "Influence of diatom diversity on the ocean biological carbon pump". Nature Geoscience 11 (1): 27–37. doi:10.1038/s41561-017-0028-x. Bibcode: 2018NatGe..11...27T. https://hal.science/hal-01667978/file/treguer2017.pdf.
- ↑ Mahadevan, Amala; d'Asaro, Eric; Lee, Craig; Perry, Mary Jane (2012). "Eddy-Driven Stratification Initiates North Atlantic Spring Phytoplankton Blooms". Science 337 (6090): 54–58. doi:10.1126/science.1218740. PMID 22767922. Bibcode: 2012Sci...337...54M.
- ↑ 62.0 62.1 62.2 62.3 Cavicchioli, Ricardo; Ripple, William J.; Timmis, Kenneth N.; Azam, Farooq; Bakken, Lars R.; Baylis, Matthew; Behrenfeld, Michael J.; Boetius, Antje et al. (2019). "Scientists' warning to humanity: Microorganisms and climate change". Nature Reviews Microbiology 17 (9): 569–586. doi:10.1038/s41579-019-0222-5. PMID 31213707.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Wassilieff, Maggy (2006) "Plankton - Plant plankton", Te Ara - the Encyclopedia of New Zealand. Accessed: 2 November 2019.
- ↑ "King's College London - Lake Megachad" (in en-GB). https://www.kcl.ac.uk/sspp/departments/geography/people/academic/drake/Research/The-Sahara-Megalakes-Project/Lake-Megachad.aspx.
- ↑ Boyd, Philip W.; Claustre, Hervé; Levy, Marina; Siegel, David A.; Weber, Thomas (2019). "Multi-faceted particle pumps drive carbon sequestration in the ocean". Nature 568 (7752): 327–335. doi:10.1038/s41586-019-1098-2. PMID 30996317. Bibcode: 2019Natur.568..327B. https://hal.archives-ouvertes.fr/hal-02117441/file/pdf%20merged%20of%20final%20submission%20317960_6_merged_1547169923.pdf.
- ↑ Zhang, D.; Wang, Y.; Cai, J.; Pan, J.; Jiang, X.; Jiang, Y. (2012). "Bio-manufacturing technology based on diatom micro- and nanostructure". Chinese Science Bulletin 57 (30): 3836–3849. doi:10.1007/s11434-012-5410-x. Bibcode: 2012ChSBu..57.3836Z.
- ↑ Behrenfeld, Michael J.; Doney, Scott C.; Lima, Ivan; Boss, Emmanuel S.; Siegel, David A. (2013). "Annual cycles of ecological disturbance and recovery underlying the subarctic Atlantic spring plankton bloom". Global Biogeochemical Cycles 27 (2): 526–540. doi:10.1002/gbc.20050. Bibcode: 2013GBioC..27..526B.
- ↑ Rousseaux, Cecile S.; Gregg, Watson W. (2015). "Recent decadal trends in global phytoplankton composition". Global Biogeochemical Cycles 29 (10): 1674–1688. doi:10.1002/2015GB005139. Bibcode: 2015GBioC..29.1674R.
- ↑ Arsenieff, L.; Simon, N.; Rigaut-Jalabert, F.; Le Gall, F.; Chaffron, S.; Corre, E.; Com, E.; Bigeard, E. et al. (2018). "First Viruses Infecting the Marine Diatom Guinardia delicatula". Frontiers in Microbiology 9: 3235. doi:10.3389/fmicb.2018.03235. PMID 30687251.
- ↑ Kilias, Estelle S.; Junges, Leandro; Šupraha, Luka; Leonard, Guy; Metfies, Katja; Richards, Thomas A. (2020). "Chytrid fungi distribution and co-occurrence with diatoms correlate with sea ice melt in the Arctic Ocean". Communications Biology 3 (1): 183. doi:10.1038/s42003-020-0891-7. PMID 32317738.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Rost, B. and Riebesell, U. (2004) "Coccolithophores and the biological pump: responses to environmental changes". In: Coccolithophores: From Molecular Processes to Global Impact, pages 99–125, Springer. ISBN 9783662062784.
- ↑ 72.0 72.1 Wassilieff, Maggy (2006) "A coccolithophore", Te Ara - the Encyclopedia of New Zealand. Accessed: 2 November 2019.
- ↑ Young, J.R.; Pratiwi, S.; Su, X. (2017). Data report: Surface seawater plankton sampling for coccolithophores undertaken during IODP Expedition 359. Proceedings of the International Ocean Discovery Program. doi:10.14379/iodp.proc.359.111.2017.
- ↑ Hagino, K., Onuma, R., Kawachi, M. and Horiguchi, T. (2013) "Discovery of an endosymbiotic nitrogen-fixing cyanobacterium UCYN-A in Braarudosphaera bigelowii (Prymnesiophyceae)". PLoS One, 8(12): e81749. doi:10.1371/journal.pone.0081749.
- ↑ Gómez F (2012). "A checklist and classification of living dinoflagellates (Dinoflagellata, Alveolata)". CICIMAR Oceánides 27 (1): 65–140. doi:10.37543/oceanides.v27i1.111.
- ↑ Stoecker, D. K. (1999). "Mixotrophy among Dinoflagellates". The Journal of Eukaryotic Microbiology 46 (4): 397–401. doi:10.1111/j.1550-7408.1999.tb04619.x.
- ↑ Suggested Explanation for Glowing Seas--Including Currently Glowing California Seas National Science Foundation, 18 October 2011.
- ↑ Boltovskoy, Demetrio; Anderson, O. Roger; Correa, Nancy M. (2017). "Radiolaria and Phaeodaria" (in en). Handbook of the Protists. Springer, Cham. pp. 731–763. doi:10.1007/978-3-319-28149-0_19. ISBN 9783319281476.
- ↑ Anderson, O. R. (1983). Radiolaria. Springer Science & Business Media.
- ↑ Gast, R. J.; Caron, D. A. (1996-11-01). "Molecular phylogeny of symbiotic dinoflagellates from planktonic foraminifera and radiolaria" (in en). Molecular Biology and Evolution 13 (9): 1192–1197. doi:10.1093/oxfordjournals.molbev.a025684. ISSN 0737-4038. PMID 8896371.
- ↑ Castro, Peter; Huber, Michael E. (2010). Marine Biology (8th ed.). McGraw Hill. pp. 95. ISBN 978-0071113021. https://archive.org/details/marinebiology00cast_419.
- ↑ "Chemistries and colors of bioluminescent reactions: a review". Gene 173 (1 Spec No): 5–11. 1996. doi:10.1016/0378-1119(95)00676-1. PMID 8707056.
- ↑ "Bioluminescence in the sea". Annual Review of Marine Science 2: 443–93. 2009. doi:10.1146/annurev-marine-120308-081028. PMID 21141672. Bibcode: 2010ARMS....2..443H.
- ↑ "Protozoa Infecting Gills and Skin". The Merck Veterinary Manual. http://www.merckvetmanual.com:80/mvm/index.jsp?cfile=htm/bc/170410.htm.
- ↑ Brand, Larry E.; Campbell, Lisa; Bresnan, Eileen (2012). "Karenia: The biology and ecology of a toxic genus". Harmful Algae 14: 156–178. doi:10.1016/j.hal.2011.10.020. PMID 36733478.
- ↑ Buskey, E.J. (1995). "Growth and bioluminescence of Noctiluca scintillans on varying algal diets". Journal of Plankton Research 17 (1): 29–40. doi:10.1093/plankt/17.1.29.
- ↑ Panno, Joseph (14 May 2014) (in en). The Cell: Evolution of the First Organism. Infobase Publishing. ISBN 9780816067367. https://books.google.com/books?id=sYgKY6zz20YC&q=panno+the+cell&pg=PA130.
- ↑ Bertrand, Jean-Claude; Caumette, Pierre; Lebaron, Philippe; Matheron, Robert; Normand, Philippe; Sime-Ngando, Télesphore (2015-01-26) (in en). Environmental Microbiology: Fundamentals and Applications: Microbial Ecology. Springer. ISBN 9789401791182. https://books.google.com/books?id=2zVqBgAAQBAJ&q=endocytosis&pg=PA9.
- ↑ Madigan, Michael T. (2012). Brock Biology of Microorganisms. Benjamin Cummings. ISBN 9780321649638. https://books.google.com/books?id=RawZTwEACAAJ&q=brock+biology+of+microorganisms+13th.
- ↑ Yaeger, Robert G. (1996). Protozoa: Structure, Classification, Growth, and Development. NCBI. ISBN 9780963117212. https://www.ncbi.nlm.nih.gov/books/NBK8325/. Retrieved 2018-03-23.
- ↑ 91.0 91.1 Wassilieff, Maggy (2006) "Plankton - Animal plankton", Te Ara - the Encyclopedia of New Zealand. Accessed: 2 November 2019.
- ↑ Varea, C.; Aragon, J.L.; Barrio, R.A. (1999). "Turing patterns on a sphere". Physical Review E 60 (4): 4588–92. doi:10.1103/PhysRevE.60.4588. PMID 11970318. Bibcode: 1999PhRvE..60.4588V.
- ↑ 93.0 93.1 Hemleben, C.; Anderson, O.R.; Spindler, M. (1989). Modern Planktonic Foraminifera. Springer-Verlag. ISBN 978-3-540-96815-3. https://books.google.com/books?id=NaHOmAEACAAJ.
- ↑ Foraminifera: History of Study, University College London. Retrieved: 18 November 2019.
- ↑ Advances in Microbial Ecology, Volum 11
- ↑ Bernhard, J. M.; Bowser, S.M. (1999). "Benthic Foraminifera of dysoxic sediments: chloroplast sequestration and functional morphology". Earth-Science Reviews 46 (1): 149–165. doi:10.1016/S0012-8252(99)00017-3. Bibcode: 1999ESRv...46..149B.
- ↑ Calbet, Albert; Landry, Michael R. (2004). "Phytoplankton growth, microzooplankton grazing, and carbon cycling in marine systems". Limnology and Oceanography 49 (1): 51–57. doi:10.4319/lo.2004.49.1.0051. Bibcode: 2004LimOc..49...51C.
- ↑ 98.0 98.1 98.2 98.3 Haraguchi, Lumi; Jakobsen, Hans H.; Lundholm, Nina; Carstensen, Jacob (2018). "Phytoplankton Community Dynamic: A Driver for Ciliate Trophic Strategies". Frontiers in Marine Science 5. doi:10.3389/fmars.2018.00272.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Azam, F.; Fenchel, T.; Field, JG; Gray, JS; Meyer-Reil, LA; Thingstad, F. (1983). "The Ecological Role of Water-Column Microbes in the Sea". Marine Ecology Progress Series 10: 257–263. doi:10.3354/meps010257. Bibcode: 1983MEPS...10..257A.
- ↑ Sherr, Evelyn; Sherr, Barry (1988). "Role of microbes in pelagic food webs: A revised concept". Limnology and Oceanography 33 (5): 1225–1227. doi:10.4319/lo.1988.33.5.1225. Bibcode: 1988LimOc..33.1225S.
- ↑ Fenchel, T. (1988). "Marine Plankton Food Chains". Annual Review of Ecology and Systematics 19: 19–38. doi:10.1146/annurev.es.19.110188.000315.
- ↑ Buitenhuis, Erik; Le Quéré, Corinne; Aumont, Olivier; Beaugrand, Grégory; Bunker, Adrian; Hirst, Andrew; Ikeda, Tsutomu; O'Brien, Todd et al. (2006). "Biogeochemical fluxes through mesozooplankton". Global Biogeochemical Cycles 20 (2): n/a. doi:10.1029/2005GB002511. Bibcode: 2006GBioC..20.2003B.
- ↑ Behrenfeld, Michael J.; Falkowski, Paul G. (1997). "Photosynthetic rates derived from satellite-based chlorophyll concentration". Limnology and Oceanography 42 (1): 1–20. doi:10.4319/lo.1997.42.1.0001. Bibcode: 1997LimOc..42....1B.
- ↑ Calbet, Albert (2001). "Mesozooplankton grazing effect on primary production: A global comparative analysis in marine ecosystems". Limnology and Oceanography 46 (7): 1824–1830. doi:10.4319/lo.2001.46.7.1824. Bibcode: 2001LimOc..46.1824C.
- ↑ Landry, Michael R.; Calbet, Albert (2004). "Microzooplankton production in the oceans". ICES Journal of Marine Science 61 (4): 501–507. doi:10.1016/j.icesjms.2004.03.011.
- ↑ 106.0 106.1 Buitenhuis, Erik T.; Rivkin, Richard B.; Sailley, Sévrine; Le Quéré, Corinne (2010). "Biogeochemical fluxes through microzooplankton". Global Biogeochemical Cycles 24 (4): n/a. doi:10.1029/2009GB003601. Bibcode: 2010GBioC..24.4015B.
- ↑ Hansen, Per Juel; Bjørnsen, Peter Koefoed; Hansen, Benni Winding (2000). "Zooplankton grazing and growth: Scaling within the 2-2,000-µm body size range". Limnology and Oceanography 45 (8): 1891. doi:10.4319/lo.2000.45.8.1891. Bibcode: 2000LimOc..45.1891H.
- ↑ Nielsen, Torkel Gissel; Kicrboe, Thomas (1994). "Regulation of zooplankton biomass and production in a temperate, coastal ecosystem. 2. Ciliates". Limnology and Oceanography 39 (3): 508–519. doi:10.4319/lo.1994.39.3.0508. Bibcode: 1994LimOc..39..508N.
- ↑ Stoecker, Diane K.; Capuzzo, Judith Mcdowell (1990). "Predation on Protozoa: its importance to zooplankton". Journal of Plankton Research 12 (5): 891–908. doi:10.1093/plankt/12.5.891.
- ↑ Gifford, Dian J. (1991). "The Protozoan-Metazoan Trophic Link in Pelagic Ecosystems". The Journal of Protozoology 38: 81–86. doi:10.1111/j.1550-7408.1991.tb04806.x.
- ↑ Flynn, Kevin J.; Stoecker, Diane K.; Mitra, Aditee; Raven, John A.; Glibert, Patricia M.; Hansen, Per Juel; Granéli, Edna; Burkholder, Joann M. (2013). "Misuse of the phytoplankton–zooplankton dichotomy: The need to assign organisms as mixotrophs within plankton functional types". Journal of Plankton Research 35: 3–11. doi:10.1093/plankt/fbs062.
- ↑ Edwards, Kyle F.; Thomas, Mridul K.; Klausmeier, Christopher A.; Litchman, Elena (2012). "Allometric scaling and taxonomic variation in nutrient utilization traits and maximum growth rate of phytoplankton". Limnology and Oceanography 57 (2): 554–566. doi:10.4319/lo.2012.57.2.0554. Bibcode: 2012LimOc..57..554E.
- ↑ 113.0 113.1 Mitra, A.; Flynn, K. J.; Burkholder, J. M.; Berge, T.; Calbet, A.; Raven, J. A.; Granéli, E.; Glibert, P. M. et al. (2014). "The role of mixotrophic protists in the biological carbon pump". Biogeosciences 11 (4): 995–1005. doi:10.5194/bg-11-995-2014. Bibcode: 2014BGeo...11..995M.
- ↑ Ward, Ben A.; Follows, Michael J. (2016). "Marine mixotrophy increases trophic transfer efficiency, mean organism size, and vertical carbon flux". Proceedings of the National Academy of Sciences 113 (11): 2958–2963. doi:10.1073/pnas.1517118113. PMID 26831076. Bibcode: 2016PNAS..113.2958W.
- ↑ Leles, S. G.; Mitra, A.; Flynn, K. J.; Stoecker, D. K.; Hansen, P. J.; Calbet, A.; McManus, G. B.; Sanders, R. W. et al. (2017). "Oceanic protists with different forms of acquired phototrophy display contrasting biogeographies and abundance". Proceedings of the Royal Society B: Biological Sciences 284 (1860). doi:10.1098/rspb.2017.0664. PMID 28768886.
- ↑ Matz, Mikhail V.; Tamara M. Frank; N. Justin Marshall; Edith A. Widder; Sonke Johnsen (2008-12-09). "Giant Deep-Sea Protist Produces Bilaterian-like Traces". Current Biology (Elsevier Ltd) 18 (23): 1849–1854. doi:10.1016/j.cub.2008.10.028. PMID 19026540. http://www.biology.duke.edu/johnsenlab/pdfs/pubs/sea%20grapes%202008.pdf.
- ↑ Gooday, A. J.; Aranda da Silva, A.; Pawlowski, J. (2011-12-01). "Xenophyophores (Rhizaria, Foraminifera) from the Nazaré Canyon (Portuguese margin, NE Atlantic)". Deep-Sea Research Part II: Topical Studies in Oceanography. The Geology, Geochemistry, and Biology of Submarine Canyons West of Portugal 58 (23–24): 2401–2419. doi:10.1016/j.dsr2.2011.04.005. Bibcode: 2011DSRII..58.2401G.
- ↑ 118.00 118.01 118.02 118.03 118.04 118.05 118.06 118.07 118.08 118.09 118.10 Bjorbækmo, Marit F. Markussen; Evenstad, Andreas; Røsæg, Line Lieblein; Krabberød, Anders K.; Logares, Ramiro (2020). "The planktonic protist interactome: where do we stand after a century of research?". The ISME Journal 14 (2): 544–559. doi:10.1038/s41396-019-0542-5. PMID 31685936. 50px Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Margulis, Lynn; Fester, René (1991). Symbiosis as a Source of Evolutionary Innovation: Speciation and Morphogenesis. MIT Press. ISBN 9780262132695. https://books.google.com/books?id=3sKzeiHUIUQC&q=%22Symbiosis+as+a+source+of+evolutionary+innovation%22.
- ↑ López-García, Purificación; Eme, Laura; Moreira, David (2017). "Symbiosis in eukaryotic evolution". Journal of Theoretical Biology 434: 20–33. doi:10.1016/j.jtbi.2017.02.031. PMID 28254477. Bibcode: 2017JThBi.434...20L.
- ↑ Archibald, John M. (2015). "Endosymbiosis and Eukaryotic Cell Evolution". Current Biology 25 (19): R911–R921. doi:10.1016/j.cub.2015.07.055. PMID 26439354.
- ↑ Cavalier-Smith, Thomas (2013). "Symbiogenesis: Mechanisms, Evolutionary Consequences, and Systematic Implications". Annual Review of Ecology, Evolution, and Systematics 44: 145–172. doi:10.1146/annurev-ecolsys-110411-160320.
- ↑ Mahé, Frédéric; De Vargas, Colomban; Bass, David; Czech, Lucas; Stamatakis, Alexandros; Lara, Enrique; Singer, David; Mayor, Jordan et al. (2017). "Parasites dominate hyperdiverse soil protist communities in Neotropical rainforests". Nature Ecology & Evolution 1 (4): 91. doi:10.1038/s41559-017-0091. PMID 28812652.
- ↑ Biard, Tristan; Stemmann, Lars; Picheral, Marc; Mayot, Nicolas; Vandromme, Pieter; Hauss, Helena; Gorsky, Gabriel; Guidi, Lionel et al. (2016). "In situ imaging reveals the biomass of giant protists in the global ocean". Nature 532 (7600): 504–507. doi:10.1038/nature17652. PMID 27096373. Bibcode: 2016Natur.532..504B. https://hal.sorbonne-universite.fr/hal-01324873/file/Biard_2016_In_situ_imaging.pdf.
- ↑ Finlay, B.J.; Esteban, G.F. (1998). "Freshwater protozoa: Biodiversity and ecological function". Biodiversity and Conservation 7 (9): 1163–1186. doi:10.1023/A:1008879616066.
- ↑ Huxley, Thomas H. (1851). "XXXIV.—Zoological notes and observations made on board H.M.S. Rattlesnake". Annals and Magazine of Natural History 8 (48): 433–442. doi:10.1080/03745486109495002. https://zenodo.org/record/2454610.
- ↑ Brandt K. (1881) "Uber das Zusammenleben von Thieren und Algen". Verh Physiol Ges, 1: 524–527.
- ↑ Logares, Ramiro; Haverkamp, Thomas H.A.; Kumar, Surendra; Lanzén, Anders; Nederbragt, Alexander J.; Quince, Christopher; Kauserud, Håvard (2012). "Environmental microbiology through the lens of high-throughput DNA sequencing: Synopsis of current platforms and bioinformatics approaches". Journal of Microbiological Methods 91 (1): 106–113. doi:10.1016/j.mimet.2012.07.017. PMID 22849829.
- ↑ Sogin, M. L.; Morrison, H. G.; Huber, J. A.; Welch, D. M.; Huse, S. M.; Neal, P. R.; Arrieta, J. M.; Herndl, G. J. (2006). "Microbial diversity in the deep sea and the underexplored "rare biosphere"". Proceedings of the National Academy of Sciences 103 (32): 12115–12120. doi:10.1073/pnas.0605127103. PMID 16880384. Bibcode: 2006PNAS..10312115S.
- ↑ Goodwin, Sara; McPherson, John D.; McCombie, W. Richard (2016). "Coming of age: Ten years of next-generation sequencing technologies". Nature Reviews Genetics 17 (6): 333–351. doi:10.1038/nrg.2016.49. PMID 27184599.
- ↑ Pedrós-Alió C, Acinas SG, Logares R, Massana R. Marine microbial diversity as seen by high throughput sequencing. In: Gasol, Josep M.; Kirchman, David L. (27 March 2018). Microbial Ecology of the Oceans. John Wiley & Sons. ISBN 9781119107187. https://books.google.com/books?id=fKJFDwAAQBAJ&q=%22Microbial+ecology+of+the+oceans%22., pp. 47–87.
- ↑ Spang, Anja; Saw, Jimmy H.; Jørgensen, Steffen L.; Zaremba-Niedzwiedzka, Katarzyna; Martijn, Joran; Lind, Anders E.; Van Eijk, Roel; Schleper, Christa et al. (2015). "Complex archaea that bridge the gap between prokaryotes and eukaryotes". Nature 521 (7551): 173–179. doi:10.1038/nature14447. PMID 25945739. Bibcode: 2015Natur.521..173S.
- ↑ Faust, Karoline; Lahti, Leo; Gonze, Didier; De Vos, Willem M.; Raes, Jeroen (2015). "Metagenomics meets time series analysis: Unraveling microbial community dynamics". Current Opinion in Microbiology 25: 56–66. doi:10.1016/j.mib.2015.04.004. PMID 26005845.
- ↑ Faust, Karoline; Raes, Jeroen (2012). "Microbial interactions: From networks to models". Nature Reviews Microbiology 10 (8): 538–550. doi:10.1038/nrmicro2832. PMID 22796884.
- ↑ Lima-Mendez, G. et al. (2015). "Determinants of community structure in the global plankton interactome". Science 348 (6237). doi:10.1126/science.1262073. PMID 25999517.
- ↑ Layeghifard, Mehdi; Hwang, David M.; Guttman, David S. (2017). "Disentangling Interactions in the Microbiome: A Network Perspective". Trends in Microbiology 25 (3): 217–228. doi:10.1016/j.tim.2016.11.008. PMID 27916383.
- ↑ Adl, Sina M. et al. (2019). "Revisions to the classification, nomenclature, and diversity of eukaryotes". Journal of Eukaryotic Microbiology 66 (1): 4–119. doi:10.1111/jeu.12691. PMID 30257078.
- ↑ Schulz, Frederik; Eloe-Fadrosh, Emiley A.; Bowers, Robert M.; Jarett, Jessica; Nielsen, Torben; Ivanova, Natalia N.; Kyrpides, Nikos C.; Woyke, Tanja (2017). "Towards a balanced view of the bacterial tree of life". Microbiome 5 (1): 140. doi:10.1186/s40168-017-0360-9. PMID 29041958.
- ↑ "Groups of Protists | Boundless Biology". https://courses.lumenlearning.com/boundless-biology/chapter/groups-of-protists/.
- ↑ 140.0 140.1 Aguirre, L.E., Ouyang, L., Elfwing, A., Hedblom, M., Wulff, A. and Inganäs, O. (2018) "Diatom frustules protect DNA from ultraviolet light". Scientific reports, 8(1): 1–6. doi:10.1038/s41598-018-21810-2.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ De Tommasi, E., Congestri, R., Dardano, P., De Luca, A.C., Managò, S., Rea, I. and De Stefano, M. (2018) "UV-shielding and wavelength conversion by centric diatom nanopatterned frustules". Scientific Reports, 8(1): 1–14. doi:10.1038/s41598-018-34651-w.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ Biodegradable glitter and pollution-eating microalgae: the new materials inspired by nature Horizon, 28 May 2020.
- ↑ Gafar, N.A., Eyre, B.D. and Schulz, K.G. (2019) "A comparison of species specific sensitivities to changing light and carbonate chemistry in calcifying marine phytoplankton". Scientific Reports, 9(1): 1–12. doi:10.1038/s41598-019-38661-0.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
- ↑ 144.0 144.1 144.2 144.3 Monteiro, F.M., Bach, L.T., Brownlee, C., Bown, P., Rickaby, R.E., Poulton, A.J., Tyrrell, T., Beaufort, L., Dutkiewicz, S., Gibbs, S. and Gutowska, M.A. (2016) "Why marine phytoplankton calcify". Science Advances, 2(7): e1501822. doi:10.1126/sciadv.1501822.
 Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Modified text was copied from this source, which is available under a Creative Commons Attribution 4.0 International License.
Further references
- Bjorbækmo, Marit F. Markussen; Evenstad, Andreas; Røsæg, Line Lieblein; Krabberød, Anders K.; Logares, Ramiro (2020). "The planktonic protist interactome: Where do we stand after a century of research?". The ISME Journal 14 (2): 544–559. doi:10.1038/s41396-019-0542-5. PMID 31685936.
 Available under a Creative Commons Attribution 4.0 International License.
Available under a Creative Commons Attribution 4.0 International License. - Flynn, Kevin J.; Mitra, Aditee; Anestis, Konstantinos; Anschütz, Anna A.; Calbet, Albert; Ferreira, Guilherme Duarte; Gypens, Nathalie; Hansen, Per J. et al. (2019). "Mixotrophic protists and a new paradigm for marine ecology: Where does plankton research go now?". Journal of Plankton Research 41 (4): 375–391. doi:10.1093/plankt/fbz026.
- Leles, Suzana Gonçalves; Polimene, Luca; Bruggeman, Jorn; Blackford, Jeremy; Ciavatta, Stefano; Mitra, Aditee; Flynn, Kevin John (2018). "Modelling mixotrophic functional diversity and implications for ecosystem function". Journal of Plankton Research 40 (6): 627–642. doi:10.1093/plankt/fby044.
- Keeling, Patrick J.; Campo, Javier del (2017). "Marine Protists Are Not Just Big Bacteria". Current Biology (Elsevier BV) 27 (11): R541–R549. doi:10.1016/j.cub.2017.03.075. ISSN 0960-9822. PMID 28586691.
 |