Biology:Middle East respiratory syndrome–related coronavirus
| Middle East respiratory syndrome–related coronavirus | |
|---|---|
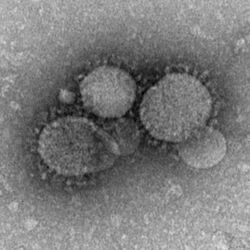
| |
| MERS-CoV particles as seen by negative stain electron microscopy. Virions contain characteristic club-like projections emanating from the viral membrane. | |
| Virus classification | |
| (unranked): | Virus |
| Realm: | Riboviria |
| Kingdom: | Orthornavirae |
| Phylum: | Pisuviricota |
| Class: | Pisoniviricetes |
| Order: | Nidovirales |
| Family: | Coronaviridae |
| Genus: | Betacoronavirus |
| Subgenus: | Merbecovirus |
| Species: | Middle East respiratory syndrome–related coronavirus
|
Middle East respiratory syndrome–related coronavirus (MERS-CoV),[1] or EMC/2012 (HCoV-EMC/2012), is the virus that causes Middle East respiratory syndrome (MERS).[2][3] It is a species of coronavirus which infects humans, bats, and camels.[4] The infecting virus is an enveloped, positive-sense, single-stranded RNA virus which enters its host cell by binding to the DPP4 receptor.[5] The species is a member of the genus Betacoronavirus and subgenus Merbecovirus.[6][4]
Initially called simply novel coronavirus or nCoV, it was first reported in June 2012 after genome sequencing of a virus isolated from sputum samples from a person who fell ill in a 2012 outbreak of a new flu-like respiratory illness. By July 2015, MERS-CoV cases had been reported in over 21 countries, in Europe, North America and Asia as well as the Middle East. MERS-CoV is one of several viruses identified by the World Health Organization (WHO) as a likely cause of a future epidemic. They list it for urgent research and development.[7][8]
Virology
The virus MERS-CoV is a member of the beta group of coronavirus, Betacoronavirus, lineage C. MERS-CoV genomes are phylogenetically classified into two clades, clade A and B. The earliest cases were of clade A clusters, while the majority of more recent cases are of the genetically distinct clade B.[9]
MERS-CoV is one of seven known coronaviruses to infect humans, including HCoV-229E, HCoV-NL63, HCoV-OC43, HCoV-HKU1, the original SARS-CoV (or SARS-CoV-1), and SARS-CoV-2.[10] It has frequently been referred to as a SARS-like virus.[11] By November, 2019, 2,494 cases of MERS had been reported with 858 deaths, implying a case fatality rate of greater than 30%.[12]
Early cases and spillover event
The first confirmed case was reported in Jeddah, Saudi Arabia in April 2012.[10] Egyptian virologist Ali Mohamed Zaki isolated and identified a previously unknown coronavirus from the man's lungs.[13][14][15] Zaki then posted his findings on 24 September 2012 on ProMED-mail.[14][16] The isolated cells showed cytopathic effects (CPE), in the form of rounding and syncytia formation.[16]
A second case was found in September 2012, when a 49-year-old man living in Qatar presented with similar flu symptoms. A sequence of the virus was nearly identical to that of the first case.[10] In November 2012, similar cases appeared in Qatar and Saudi Arabia. Additional cases were noted, with deaths associated, and rapid research and monitoring of the novel coronavirus began. It is not known whether the infections are the result of a single zoonotic event with subsequent human-to-human transmission, or if the multiple geographic sites of infection represent multiple zoonotic events from an unknown common source.[citation needed]
A study by Ziad Memish of Riyadh University and colleagues suggests that the virus arose some time between July 2007 and June 2012, with perhaps as many as seven separate zoonotic transmissions.[citation needed] Among animal reservoirs, CoV has a large genetic diversity yet the samples from patients suggested a similar genome, and therefore common source, though the data were limited. It was determined through molecular clock analysis that viruses from the EMC/2012 and England/Qatar/2012 date to early 2011, suggesting that these cases were descended from a single zoonotic event. It appeared the MERS-CoV had been circulating in the human population for more than a year without detection, and suggested independent transmission from an unknown source.[17][18]
Tropism
In humans, the virus has a strong tropism for nonciliated bronchial epithelial cells, and it has been shown to effectively evade the innate immune responses and antagonize interferon (IFN) production in these cells. This tropism is unique in that most respiratory viruses target ciliated cells.[19][20]
Due to the clinical similarity between MERS-CoV and SARS-CoV, it was proposed that they may use the same cellular receptor; the exopeptidase, angiotensin converting enzyme 2 (ACE2).[21] However, it was later discovered that neutralization of ACE2 by recombinant antibodies does not prevent MERS-CoV infection.[22] Further research identified dipeptidyl peptidase 4 (DPP4; also known as CD26) as a functional cellular receptor for MERS-CoV.[20] Unlike other known coronavirus receptors, the enzymatic activity of DPP4 is not required for infection. As would be expected, the amino acid sequence of DPP4 is highly conserved across species and is expressed in the human bronchial epithelium and kidneys.[20][23] Bat DPP4 genes appear to have been subject to a high degree of adaptive evolution as a response to coronavirus infections, so the lineage leading to MERS-CoV may have circulated in bat populations for a long period of time before being transmitted to people.[24]
Transmission
On 13 February 2013, the World Health Organization stated that "the risk of sustained person-to-person transmission appears to be very low."[25] The cells MERS-CoV infects in the lungs only account for 20% of respiratory epithelial cells, so a large number of virions are likely needed to be inhaled to cause infection.[23]
Anthony Fauci of the National Institutes of Health in Bethesda, Maryland, stated that MERS-CoV "does not spread in a sustained person to person way at all," while noting the possibility that the virus could mutate into a strain that does transmit from person to person.[26] However, the infection of healthcare workers has led to concerns of human to human transmission.[27]
The Centers for Disease Control and Prevention (CDC) list MERS as transmissible from human to human.[28] They state that "MERS-CoV has been shown to spread between people who are in close contact. Transmission from infected patients to healthcare personnel has also been observed. Clusters of cases in several countries are being investigated."[28][29]
However, on the 28th of May, the CDC revealed that the Illinois man who was originally thought to have been the first incidence of person-to-person spread (from the Indiana man at a business meeting), had in fact tested negative for MERS-CoV. After completing additional and more definitive tests using a neutralising antibody assay, experts at the CDC concluded that the Indiana patient did not spread the virus to the Illinois patient. Tests concluded that the Illinois man had not been previously infected. It is possible for MERS to be symptomless, and early research has shown that up to 20% of cases show no signs of active infection but have MERS-CoV antibodies in their blood.[30]
Evolution
The virus appears to have originated in bats.[31] The virus itself has been isolated from a bat.[32] This virus is closely related to the Tylonycteris bat coronavirus HKU4 and Pipistrellus bat coronavirus HKU5.[33] Serological evidence shows that these viruses have infected camels for at least 20 years. The most recent common ancestor of several human strains has been dated to March 2012 (95% confidence interval December 2011 to June 2012).[34]
It is thought that the viruses have been present in bats for some time and had spread to camels by the mid-1990s. The viruses appear to have spread from camels to humans in the early 2010s. The original bat host species and the time of initial infection in this species has yet to be determined.[citation needed] Examination of the sequences of 238 isolates suggested that this virus has evolved into three clades differing in codon usage, host, and geographic distribution.[35]
Natural reservoir
Early research suggested the virus is related to one found in the Egyptian tomb bat.[citation needed] It is believed that the virus originated in bats.[36] Work by epidemiologist Ian Lipkin of Columbia University in New York showed that the virus isolated from a bat looked to be a match to the virus found in humans.[37][38][39] 2c betacoronaviruses were detected in Nycteris bats in Ghana and Pipistrellus bats in Europe that are phylogenetically related to the MERS-CoV virus.[40] However the major natural reservoir where humans get the virus infection remained unknown until on 9 August 2013, a report in the journal The Lancet Infectious Diseases showed that 50 out of 50 (100%) blood serum from Omani camels and 15 of 105 (14%) from Spanish camels had protein-specific antibodies against the MERS-CoV spike protein. Blood serum from European sheep, goats, cattle, and other camelids had no such antibodies.[41]
Soon after on 5 September 2013 a seroepidemiological study published in the journal of Eurosurveillance by R.A Perera et al.[42] where they investigated 1343 human and 625 animal sera indicated, the abundant presence of MERS-CoV specific antibody in 108 out of 110 Egyptian dromedary camels but not in other animals such as goats, cows or sheep in this region.[42] These are the first and significant scientific reports that indicated the role of "dromedary camels" as a reservoir of MERS-CoV.
Research has linked camels, showing that the coronavirus infection in dromedary camel calves and adults is a 99.9% match to the genomes of human clade B MERS-CoV.[43] At least one person who has fallen sick with MERS was known to have come into contact with camels or recently drank camel milk.[44] Countries like Saudi Arabia and the United Arab Emirates produce and consume large amounts of camel meat. The possibility exists that African or Australian bats harbor the virus and transmit it to camels. Imported camels from these regions might have carried the virus to the Middle East.[45]
In 2013 MERS-CoV was identified in three members of a dromedary camel herd held in a Qatar barn, which was linked to two confirmed human cases who have since recovered. The presence of MERS-CoV in the camels was confirmed by the National Institute of Public Health and Environment (RIVM) of the Ministry of Health and the Erasmus Medical Center (WHO Collaborating Center), the Netherlands. None of the camels showed any sign of disease when the samples were collected. The Qatar Supreme Council of Health advised in November 2013 that people with underlying health conditions, such as heart disease, diabetes, kidney disease, respiratory disease, the immunosuppressed, and the elderly, avoid any close animal contacts when visiting farms and markets, and to practice good hygiene, such as washing hands.[46]
A further study on dromedary camels from Saudi Arabia published in December 2013 revealed the presence of MERS-CoV in 90% of the evaluated dromedary camels (310), suggesting that dromedary camels not only could be the main reservoir of MERS-CoV, but also the animal source of MERS.[47]
According to the 27 March 2014 MERS-CoV summary update, recent studies support that camels serve as the primary source of the MERS-CoV infecting humans, while bats may be the ultimate reservoir of the virus. Evidence includes the frequency with which the virus has been found in camels to which human cases have been exposed, seriological data which shows widespread transmission in camels, and the similarity of the camel CoV to the human CoV.[48]
On 6 June 2014, the Arab News newspaper highlighted the latest research findings in the New England Journal of Medicine in which a 44-year-old Saudi man who kept a herd of nine camels died of MERS in November 2013. His friends said they witnessed him applying a topical medicine to the nose of one of his ill camels—four of them reportedly sick with nasal discharge—seven days before he himself became stricken with MERS. Researchers sequenced the virus found in one of the sick camels and the virus that killed the man, and found that their genomes were identical. In that same article, the Arab News reported that as of 6 June 2014, there have been 689 cases of MERS reported within the Kingdom of Saudi Arabia with 283 deaths.[49]
Taxonomy
MERS-CoV is more closely related to the bat coronaviruses HKU4 and HKU5 (lineage 2C) than it is to SARS-CoV (lineage 2B) (2, 9), sharing more than 90% sequence identity with their closest relationships, bat coronaviruses HKU4 and HKU5 and therefore considered to belong to the same species by the International Committee on Taxonomy of Viruses (ICTV).[citation needed]
- Mnemonic:
- Taxon identifier:
- Scientific name: Middle East respiratory syndrome coronavirus[1]
- Common name: MERS-CoV
- Synonym: Severe acute respiratory syndrome coronavirus
- Other names:
- novel coronavirus (nCoV)
- London1 novel CoV/2012[50]
- Human Coronavirus Erasmus Medical Center/2012 (HCoV-EMC/2012)
- Rank:
- Lineage:
- › Viruses
- › ssRNA viruses
- › Group: IV; positive-sense, single-stranded RNA viruses
- › Order: Nidovirales
- › Family: Coronaviridae
- › Subfamily: Coronavirinae
- › Genus: Betacoronavirus[51]
- › Species: Betacoronavirus 1 (commonly called Human coronavirus OC43), Human coronavirus HKU1, Murine coronavirus, Pipistrellus bat coronavirus HKU5, Rousettus bat coronavirus HKU9, Severe acute respiratory syndrome-related coronavirus, Tylonycteris bat coronavirus HKU4, MERS-CoV
- › Genus: Betacoronavirus[51]
- › Subfamily: Coronavirinae
- › Family: Coronaviridae
- › Order: Nidovirales
- › Group: IV; positive-sense, single-stranded RNA viruses
- › ssRNA viruses
Strains:
- Isolate:
- Isolate:
- NCBI
Research and patent
Saudi officials had not given permission for Dr. Zaki, the first isolator of the human strain, to send a sample of the virus to Fouchier and were angered when Fouchier claimed the patent on the full genetic sequence of MERS-CoV.[54]
The editor of The Economist observed, "Concern over security must not slow urgent work. Studying a deadly virus is risky. Not studying it is riskier."[54] Dr. Zaki was fired from his job at the hospital as a result of bypassing the Saudi Ministry of Health in his announcement and sharing his sample and findings.[55][56][57][58]
At their annual meeting of the World Health Assembly in May 2013, WHO chief Margaret Chan declared that intellectual property, or patents on strains of new virus, should not impede nations from protecting their citizens by limiting scientific investigations. Deputy Health Minister Ziad Memish raised concerns that scientists who held the patent for MERS-CoV would not allow other scientists to use patented material and were therefore delaying the development of diagnostic tests.[59] Erasmus MC responded that the patent application did not restrict public health research into MERS and MERS-CoV,[60] and that the virus and diagnostic tests were shipped—free of charge—to all that requested such reagents.
Mapping
There are a number of mapping efforts focused on tracking MERS coronavirus. On 2 May 2014, the Corona Map[61] was launched to track the MERS coronavirus in realtime on the world map. The data is officially reported by WHO or the Ministry of Health of the respective country.[62] HealthMap also tracks case reports with inclusion of news and social media as data sources as part of HealthMap MERS.[63] South Korea was infected in mid-2015, with 38 deaths among 186 cases of infection.[citation needed]
See also
- European Centre for Disease Prevention and Control
- Novel virus
- Super-spreader
- Virulence
- Coalition for Epidemic Preparedness Innovations
References
- ↑ 1.0 1.1 "Middle East respiratory syndrome coronavirus (MERS-CoV): announcement of the Coronavirus Study Group". Journal of Virology 87 (14): 7790–2. July 2013. doi:10.1128/JVI.01244-13. PMID 23678167.
- ↑ "Middle East respiratory syndrome coronavirus (MERS-CoV)" (in en). https://www.who.int/en/news-room/fact-sheets/detail/middle-east-respiratory-syndrome-coronavirus-(mers-cov).
- ↑ "Middle East respiratory syndrome". Lancet 386 (9997): 995–1007. September 2015. doi:10.1016/S0140-6736(15)60454-8. PMID 26049252.
- ↑ 4.0 4.1 Wong, Antonio C. P.; Li, Xin; Lau, Susanna K. P.; Woo, Patrick C. Y. (2019-02-20). "Global Epidemiology of Bat Coronaviruses". Viruses 11 (2): 174. doi:10.3390/v11020174. PMID 30791586. "See Figure 3.".
- ↑ "Coronaviruses: an overview of their replication and pathogenesis". Coronaviruses. Methods in Molecular Biology. 1282. Springer. 2015. pp. 1–23. doi:10.1007/978-1-4939-2438-7_1. ISBN 978-1-4939-2438-7. "See Table 1."
- ↑ "Virus Taxonomy: 2018 Release". October 2018. https://talk.ictvonline.org/taxonomy/.
- ↑ Kieny, Marie-Paule. "After Ebola, a Blueprint Emerges to Jump-Start R&D". https://blogs.scientificamerican.com/guest-blog/after-ebola-a-blueprint-emerges-to-jump-start-r-d/.
- ↑ "LIST OF PATHOGENS". https://www.who.int/csr/research-and-development/list_of_pathogens/en/.
- ↑ Chu, Daniel K.W.; Poon, Leo L.M.; Gomaa, Mokhtar M.; Shehata, Mahmoud M.; Perera, Ranawaka A.P.M.; Abu Zeid, Dina; El Rifay, Amira S.; Siu, Lewis Y. et al. (June 2014). "MERS Coronaviruses in Dromedary Camels, Egypt". Emerging Infectious Diseases 20 (6): 1049–1053. doi:10.3201/eid2006.140299. PMID 24856660.
- ↑ 10.0 10.1 10.2 "ECDC Rapid Risk Assessment - Severe respiratory disease associated with a novel coronavirus". 19 Feb 2013. http://www.ecdc.europa.eu/en/publications/Publications/novel-coronavirus-rapid-risk-assessment-update.pdf.
- ↑ Saey, Tina Hesman (2013). "Story one: Scientists race to understand deadly new virus: SARS-like infection causes severe illness, but may not spread quickly among people". Science News 183 (6): 5–6. doi:10.1002/scin.5591830603. PMID 32327842.
- ↑ "Middle East Respiratory Syndrome Coronavirus MERS-CoV". WHO. November 2019. https://www.who.int/emergencies/mers-cov/en/.
- ↑ "Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia". The New England Journal of Medicine 367 (19): 1814–20. November 2012. doi:10.1056/NEJMoa1211721. PMID 23075143.
- ↑ 14.0 14.1 Falco, Miriam (24 September 2012). "New SARS-like virus poses medical mystery". CNN. http://thechart.blogs.cnn.com/2012/09/24/new-sars-like-virus-poses-medical-mystery/.
- ↑ Dziadosz, Alexander (13 May 2013). "The doctor who discovered a new SARS-like virus says it will probably trigger an epidemic at some point, but not necessarily in its current form.". Reuters. https://www.reuters.com/article/us-coronavirus-egypt-idUSBRE94C0MH20130513.
- ↑ 16.0 16.1 "See Also". ProMED-mail. 2012-09-20. http://www.promedmail.org/direct.php?id=20120920.1302733.
- ↑ Cotten, Matthew; Lam, Tommy T.; Watson, Simon J.; Palser, Anne L.; Petrova, Velislava; Grant, Paul; Pybus, Oliver G.; Rambaut, Andrew et al. (2013-05-19). "Full-Genome Deep Sequencing and Phylogenetic Analysis of Novel Human Betacoronavirus - Vol. 19 No. 5 - May 2013 - CDC". Emerging Infectious Diseases 19 (5): 736–42B. doi:10.3201/eid1905.130057. PMID 23693015.
- ↑ "Molecular epidemiology of human coronavirus OC43 reveals evolution of different genotypes over time and recent emergence of a novel genotype due to natural recombination". Journal of Virology 85 (21): 11325–37. November 2011. doi:10.1128/JVI.05512-11. PMID 21849456.
- ↑ "Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential". mBio 4 (1): e00611–12. February 2013. doi:10.1128/mBio.00611-12. PMID 23422412.
- ↑ 20.0 20.1 20.2 "Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC". Nature 495 (7440): 251–4. March 2013. doi:10.1038/nature12005. PMID 23486063. Bibcode: 2013Natur.495..251R.
- ↑ "ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia". Journal of Virology 79 (23): 14614–21. December 2005. doi:10.1128/JVI.79.23.14614-14621.2005. PMID 16282461.
- ↑ "Human coronavirus EMC does not require the SARS-coronavirus receptor and maintains broad replicative capability in mammalian cell lines". mBio 3 (6). December 2012. doi:10.1128/mBio.00515-12. PMID 23232719.
- ↑ 23.0 23.1 Butler, Declan (2013-03-13). "Receptor for new coronavirus found". Nature 495 (7440): 149–150. doi:10.1038/495149a. PMID 23486032. Bibcode: 2013Natur.495..149B.
- ↑ "Adaptive evolution of bat dipeptidyl peptidase 4 (dpp4): implications for the origin and emergence of Middle East respiratory syndrome coronavirus". Virology Journal 10: 304. October 2013. doi:10.1186/1743-422X-10-304. PMID 24107353.
- ↑ WHO: Novel coronavirus infection – update (13 February 2013) (accessed 13 February 2013)
- ↑ "Fauci: New Virus Not Yet a 'threat to the world' (video)". Washington Times. 2012-08-31. http://www.washingtontimes.com/video/national-news/fauci-new-virus-not-yet-a-threat-to-the-world/.
- ↑ Knickmeyer, Ellen; Al Omran, Ahmed (20 Apr 2014). "Concerns Spread as New Saudi MERS Cases Spike". Wall Street Journal. https://blogs.wsj.com/middleeast/2014/04/20/concerns-spread-as-new-saudi-mers-cases-spike/.
- ↑ 28.0 28.1 "MERS-CoV - Frequently Asked Questions and Answers - Coronavirus". U.S. Centers for Disease Control and Prevention. 14 September 2017. https://www.cdc.gov/coronavirus/mers/faq.html.
- ↑ Grady, Denise (19 June 2013). "Investigation Follows Trail of a Virus in Hospitals". The New York Times. https://www.nytimes.com/2013/06/20/health/fast-spreading-virus-under-inquiry-in-saudi-arabia.html.
- ↑ Aleccia, Jonrl (28 May 2014). "CDC Backtracks: Illinois Man Didn't Have MERS After All". http://www.nbcnews.com/storyline/mers-mystery/cdc-backtracks-illinois-man-didnt-have-mers-after-all-n116436.
- ↑ "Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat". Journal of Virology 88 (19): 11297–303. October 2014. doi:10.1128/JVI.01498-14. PMID 25031349.
- ↑ "Middle East respiratory syndrome coronavirus in bats, Saudi Arabia". Emerging Infectious Diseases 19 (11): 1819–23. November 2013. doi:10.3201/eid1911.131172. PMID 24206838.
- ↑ "Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26". Cell Host & Microbe 16 (3): 328–37. September 2014. doi:10.1016/j.chom.2014.08.009. PMID 25211075.
- ↑ "Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus". mBio 5 (1): e01062–13. February 2014. doi:10.1128/mBio.01062-13. PMID 24549846.
- ↑ "Comparative Genomic Analysis MERS CoV Isolated from Humans and Camels with Special Reference to Virus Encoded Helicase". Biological & Pharmaceutical Bulletin 40 (8): 1289–1298. 2017. doi:10.1248/bpb.b17-00241. PMID 28769010.
- ↑ Mohd, Hamzah A.; Al-Tawfiq, Jaffar A.; Memish, Ziad A. (December 2016). "Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir". Virology Journal 13 (1): 87. doi:10.1186/s12985-016-0544-0.
- ↑ 37.0 37.1 Abedine, Saad (13 March 2013). "Death toll from new SARS-like virus climbs to 9". CNN. http://www.cnn.com/2013/03/13/health/new-coronavirus-case/.
- ↑ Doucleff, Michaeleen (28 September 2012). "Holy Bat Virus! Genome Hints At Origin Of SARS-Like Virus". NPR. https://www.npr.org/blogs/health/2012/09/28/161944734/holy-bat-virus-genome-hints-at-origin-of-sars-like-virus.
- ↑ jobs (2013-08-23). "Deadly coronavirus found in bats: Nature News & Comment". Nature. doi:10.1038/nature.2013.13597. http://www.nature.com/news/deadly-coronavirus-found-in-bats-1.13597. Retrieved 2014-01-19.
- ↑ 40.0 40.1 "Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe". Emerging Infectious Diseases 19 (3): 456–9. March 2013. doi:10.3201/eid1903.121503. PMID 23622767.
- ↑ "Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study". The Lancet. Infectious Diseases 13 (10): 859–66. October 2013. doi:10.1016/S1473-3099(13)70164-6. PMID 23933067.
- ↑ 42.0 42.1 Perera, R.; Wang, P.; Gomaa, M.; El-Shesheny, R.; Kandeil, A.; Bagato, O.; Siu, L.; Shehata, M. et al. (2013). "Eurosurveillance - Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013". Eurosurveillance 18 (36): 20574. doi:10.2807/1560-7917.ES2013.18.36.20574. PMID 24079378.
- ↑ "MERS coronavirus in dromedary camel herd, Saudi Arabia". Emerging Infectious Diseases 20 (7): 1231–4. July 2014. doi:10.3201/eid2007.140571. PMID 24964193.
- ↑ Roos, Robert (17 Apr 2014). "MERS outbreaks grow; Malaysian case had camel link". http://www.cidrap.umn.edu/news-perspective/2014/04/mers-outbreaks-grow-malaysian-case-had-camel-link.
- ↑ "Camels May Transmit New Middle Eastern Virus". 8 August 2013. http://news.sciencemag.org/2013/08/camels-may-transmit-new-middle-eastern-virus.
- ↑ "Three camels hit by MERS coronavirus in Qatar". Qatar Supreme Council of Health. http://www.sch.gov.qa/sch/En/catcontent.jsp?scatId=833&scatType=1&CSRT=8118031749383040885.
- ↑ "Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013". Euro Surveillance 18 (50): 20659. December 2013. doi:10.2807/1560-7917.es2013.18.50.20659. PMID 24342517.
- ↑ "Middle East respiratory syndrome coronavirus (MERS‐CoV)Summary and literature update – as of 27 March 2014". 27 March 2014. https://www.who.int/csr/disease/coronavirus_infections/MERS_CoV_Update_27_March_2014.pdf?ua=1.
- ↑ Fakeih, Mohammed Rasooldeen (6 June 2014). "80% drop in MERS infections". Arab News XXXIX (183): 1.
- ↑ Roos, Robert (25 September 2013). UK agency picks name for new coronavirus isolate (Report). University of Minnesota, Minneapolis, MN: Center for Infectious Disease Research & Policy (CIDRAP). http://www.cidrap.umn.edu/cidrap/content/other/sars/news/sep2512corona.html.
- ↑ 51.0 51.1 "Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012". Euro Surveillance 17 (40): 20290. October 2012. PMID 23078800. http://www.eurosurveillance.org/images/dynamic/EE/V17N40/art20290.pdf.
- ↑ Doucleff, Michaeleen (28 September 2012). "Holy Bat Virus! Genome Hints At Origin Of SARS-Like Virus". NPR. https://www.npr.org/blogs/health/2012/09/28/161944734/holy-bat-virus-genome-hints-at-origin-of-sars-like-virus.
- ↑ "New Coronavirus Has Many Potential Hosts, Could Pass from Animals to Humans Repeatedly". ScienceDaily. https://www.sciencedaily.com/releases/2012/12/121211083210.htm.
- ↑ 54.0 54.1 "Pandemic preparedness: Coming, ready or not". The Economist. 20 April 2013. https://www.economist.com/news/leaders/21576390-despite-progress-world-still-unprepared-new-pandemic-disease-coming-ready-or-not.
- ↑ "Egyptian Virologist Who Discovered New SARS-Like Virus Fears Its Spread". Mpelembe. 13 May 2013. http://health.mpelembe.net/home/egyptian-virologist-who-discovered-new-sars-like-virus-fears-its-spread.
- ↑ Sample, Ian; Smith, Mark (15 March 2013). "Coronavirus victim's widow tells of grief as scientists scramble for treatment". The Guardian. https://www.theguardian.com/science/2013/mar/15/coronavirus-victim-widow-scientists-treatment.
- ↑ Sample, Ian (15 March 2013). "Coronavirus: Is this the next pandemic?". The Guardian. https://www.theguardian.com/science/2013/mar/15/coronavirus-next-global-pandemic.
- ↑ Yang, Jennifer (21 October 2012). "How medical sleuths stopped a deadly new SARS-like virus in its tracks". Toronto Star. https://www.thestar.com/news/world/2012/10/21/how_medical_sleuths_stopped_a_deadly_new_sarslike_virus_in_its_tracks.html.
- ↑ "Erasmus MC: no restrictions for public health research into MERS coronavirus" (Press release). Rotterdam: Erasmus MC. 24 May 2013.
- ↑ "CoronaMap: Realtime tracking of MERS Corona virus on world map". http://CoronaMap.com/.
- ↑ "Corona Map" (Press release). 2 May 2014. Archived from the original on 4 May 2014. Retrieved 16 February 2020.
- ↑ "MERS (global map of cases)". http://healthmap.org/MERS.
External links
- Emergence of the Middle East Respiratory Syndrome Coronavirus
- MERS-CoV Complete Genome
- Emerging viruses
- Molecular Illustration of MERS-Coronavirus
- Philippines still MERS-CoV free – DOH
- Deadly Middle East Coronavirus found in an Egyptian tomb bat
Wikidata ☰ Q24808941 entry