Biology:WD40 repeat
| WD domain, G-beta repeat | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
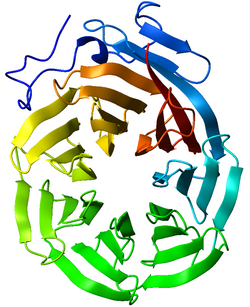 Ribbon diagram of the C-terminal WD40 domain of Tup1 (a transcriptional corepressor in yeast), which adopts a 7-bladed beta-propeller fold. Ribbon is colored from blue (N-terminus) to red (C-terminus).[1] | |||||||||||
| Identifiers | |||||||||||
| Symbol | WD40 | ||||||||||
| Pfam | PF00400 | ||||||||||
| Pfam clan | CL0186 | ||||||||||
| InterPro | IPR001680 | ||||||||||
| PROSITE | PDOC00574 | ||||||||||
| SCOP2 | 1gp2 / SCOPe / SUPFAM | ||||||||||
| CDD | cd00200 | ||||||||||
| |||||||||||
The WD40 repeat (also known as the WD or beta-transducin repeat) is a short structural motif of approximately 40 amino acids, often terminating in a tryptophan-aspartic acid (W-D) dipeptide.[2] Tandem copies of these repeats typically fold together to form a type of circular solenoid protein domain called the WD40 domain.
Structure
WD40 domain-containing proteins have 4 to 16 repeating units, all of which are thought to form a circularised beta-propeller structure (see figure to the right).[3][4] The WD40 domain is composed of several repeats, a variable region of around 20 residues at the beginning followed by a more common repeated set of residues. These repeats typically form a four stranded anti-parallel beta sheet or blade. These blades come together to form a propeller with the most common being a 7 bladed beta propeller. The blades interlock so that the last beta strand of one repeat forms with the first three of the next repeat to form the 3D blade structure.
Function
WD40-repeat proteins are a large family found in all eukaryotes and are implicated in a variety of functions ranging from signal transduction and transcription regulation to cell cycle control, autophagy and apoptosis.[5] The underlying common function of all WD40-repeat proteins is coordinating multi-protein complex assemblies, where the repeating units serve as a rigid scaffold for protein interactions. The specificity of the proteins is determined by the sequences outside the repeats themselves. Examples of such complexes are G proteins (beta subunit is a beta-propeller), TAFII transcription factor, and E3 ubiquitin ligase.[3][4]
Examples
According to the initial analysis of the human genome WD40 repeats are the eighth largest family of proteins. In all 277 proteins were identified to contain them.[6] Human genes encoding proteins containing this domain include:
- AAAS, AAMP, AHI1, AMBRA1, APAF1, ARPC1A, ARPC1B, ATG16L1,
- BOP1, BRWD1, BRWD2, BRWD3, BTRC, BUB3,
- C6orf11, CDC20, CDC40, CDRT1, CHAF1B, CIAO1, CIRH1A, COPA, COPB2, CORO1A, CORO1B, CORO1C, CORO2A, CORO2B, CORO6, CORO7, CSTF1,
- DDB2, DENND3, DMWD, DMXL1, DMXL2, DNAI1, DNAI2, DNCI1, DTL, DYNC1I1, DYNC1I2, EDC4,
- EED, EIF3S2, ELP2, EML1, EML2, EML3, EML4, EML4-ALK, EML5, ERCC8,
- FBXW10, FBXW11, FBXW2, FBXW4, FBXW5, FBXW7, FBXW8, FBXW9, FZR1,
- GBL, GEMIN5, GNB1, GNB1L, GNB2, GNB2L1, GNB3, GNB4, GNB5, GRWD1, GTF3C2,
- HERC1, HIRA, HZGJ,
- IFT121, IFT122, IFT140, IFT172, IFT80, IQWD1,
- KATNB1, KIAA1336, KIF21A, KIF21B, KM-PA-2,
- KEAP1,
- LLGL1, LLGL2, LRBA, LRRK1, LRRK2, LRWD1, LYST,
- MAPKBP1, MED16, MORG1,
- NBEA, NBEAL1, NEDD1, NLE1, NSMAF, NUP37, NUP43, NWD1,
- PAAF1, PAFAH1B1, PAK1IP1, PEX7, PHIP, PIK3R4, PLAA, PLRG1, PPP2R2A, PPP2R2B, PPP2R2C, PPP2R2D, PPWD1, PREB, PRPF19, PRPF4, PWP1, PWP2,
- RAE1, RPTOR, RBBP4, RBBP5, RBBP7, RFWD2, RFWD3, RRP9,
- SCAP, SEC13, SEC31A, SEC31B, SEH1L, SHKBP1, SMU1, SPAG16, SPG, STRAP, STRN, STRN3, STRN4, STXBP5, STXBP5L,
- TAF5, TAF5L, TBL1X, TBL1XR1, TBL1Y, TBL2, TBL3, TEP1, THOC3, THOC6, TLE1, TLE2, TLE3, TLE4, TLE6, TRAF7, TSSC1, TULP4, TUWD12,
- UTP15, UTP18,
- WAIT1, WDF3, WDFY1, WDFY2, WDFY3, WDFY4, WDHD1, WDR1, WDR10, WDR12, WDR13, WDR16, WDR17, WDR18, WDR19, WDR20, WDR21A, WDR21C, WDR22, WDR23, WDR24, WDR25, WDR26, WDR27, WDR3, WDR31, WDR32, WDR33, WDR34, WDR35, WDR36, WDR37, WDR38, WDR4, WDR40A, WDR40B, WDR40C, WDR41, WDR42A, WDR42B, WDR43, WDR44, WDR46, WDR47, WDR48, WDR49, WDR5, WDR51A, WDR51B, WDR52, WDR53, WDR54, WDR55, WDR57, WDR59, WDR5B, WDR6, WDR60, WDR61, WDR62, WDR63, WDR64, WDR65, WDR66, WDR67, WDR68, WDR69, WDR7, WDR70, WDR72, WDR73, WDR74, WDR75, WDR76, WDR77, WDR78, WDR79, WDR8, WDR81, WDR82, WDR85, WDR86, WDR88, WDR89, WDR90, WDR91, WDR92, WDSOF1, WDSUB1, WDTC1, WSB1, WSB2,
- ZFP106
| WDR gene | other gene names | NCBI Entrez Gene ID |
Human disease associated with mutations |
|---|---|---|---|
| WDR1 | AIP1; NORI-1; HEL-S-52 | 9948 | |
| WDR2 | CORO2A; IR10; CLIPINB | 7464 | |
| WDR3 | DIP2; UTP12 | 10885 | |
| WDR4 | TRM82; TRMT82 | 10785 | |
| WDR5 | SWD3; BIG-3; CFAP89 | 11091 | |
| WDR6 | 11180 | ||
| WDR7 | TRAG; KIAA0541; Rabconnectin 3 beta | 23335 | |
| WDR8 | WRAP73 | 49856 | |
| WDR9 | BRWD1; N143; C21orf107 | 54014 | |
| WDR10 | IFT122; CED; SPG; CED1; WDR10p; WDR140 | 55764 | Sensenbrenner syndrome |
| WDR11 | DR11; HH14; BRWD2; WDR15 | 55717 | Kallmann syndrome |
| WDR12 | YTM1 | 55759 | |
| WDR13 | MG21 | 64743 | |
| WDR14 | GNB1L; GY2; FKSG1; WDVCF; DGCRK3 | 54584 | |
| WDR15 | WDR11 | ||
| WDR16 | CFAP52; WDRPUH | 146845 | |
| WDR17 | 116966 | ||
| WDR18 | Ipi3 | 57418 | |
| WDR19 | ATD5; CED4; DYF-2; ORF26; Oseg6; PWDMP; SRTD5; IFT144; NPHP13 | 57728 | Sensenbrenner syndrome, Jeune syndrome |
| WDR20 | DMR | 91833 | |
| WDR21 | DCAF4; WDR21A | 26094 | |
| WDR22 | DCAF5; BCRG2; BCRP2 | 8816 | |
| WDR23 | DCAF11; GL014; PRO2389 | 80344 | |
| WDR24 | JFP7; C16orf21 | 84219 | |
| WDR25 | C14orf67 | 79446 | |
| WDR26 | CDW2; GID7; MIP2 | 80232 | |
| WDR27 | 253769 | ||
| WDR28 | GRWD1; CDW4; GRWD; RRB1 | 83743 | |
| WDR29 | SPAG16; PF20 | 79582 | |
| WDR30 | ATG16L1; IBD10; APG16L; ATG16A; ATG16L | 55054 | Crohn’s disease |
| WDR31 | 114987 | ||
| WDR32 | DCAF10 | 79269 | |
| WDR33 | NET14; WDC146 | 55339 | |
| WDR34 | DIC5; FAP133; SRTD11 | 89891 | Jeune syndrome |
| WDR35 | CED2; IFTA1; SRTD7; IFT121 | 57539 | Sensenbrenner syndrome |
| WDR36 | GLC1G; UTP21; TAWDRP; TA-WDRP | 134430 | Primary Open Angle Glaucoma |
| WDR37 | 22884 | ||
| WDR38 | 401551 | ||
| WDR39 | CIAO1; CIA1 | 9391 | |
| WDR40A | DCAF12; CT102; TCC52; KIAA1892 | 25853 | |
| WDR41 | MSTP048 | 55255 | |
| WDR43 | UTP5; NET12 | 23160 | |
| WDR44 | RPH11; RAB11BP | 54521 | |
| WDR45 | JM5; NBIA4; NBIA5; WDRX1; WIPI4; WIPI-4 | 11152 | Beta-propeller protein-associated neurodegeneration (BPAN) |
| WDR46 | UTP7; BING4; FP221; C6orf11 | 9277 | |
| WDR47 | NEMITIN; KIAA0893 | 22911 | |
| WDR48 | P80; UAF1; SPG60 | 57599 | |
| WDR49 | 151790 | ||
| WDR50 | UTP18; CGI-48 | 51096 | |
| WDR52 | CFAP44 | 55779 | |
| WDR53 | 348793 | ||
| WDR54 | 84058 | ||
| WDR55 | 54853 | ||
| WDR56 | IFT80; ATD2; SRTD2 | 57560 | Jeune syndrome |
| WDR57 | SNRNP40; SPF38; PRP8BP; HPRP8BP; PRPF8BP | 9410 | |
| WDR58 | THOC6; BBIS; fSAP35 | 79228 | |
| WDR59 | FP977 | 79726 | |
| WDR60 | SRPS6; SRTD8; FAP163 | 55112 | Jeune syndrome |
| WDR61 | SKI8; REC14 | 80349 | |
| WDR62 | MCPH2; C19orf14 | 284403 | microcephaly |
| WDR63 | DIC3; NYD-SP29 | 126820 | |
| WDR64 | 128025 | ||
| WDR65 | CFAP57; VWS2 | 149465 | Van der Woude syndrome |
| WDR66 | CaM-IP4 | 144406 | |
| WDR67 | TBC1D31; Gm85 | 93594 | |
| WDR68 | DCAF7; AN11; HAN11; SWAN-1 | 10238 | |
| WDR69 | DAW1; ODA16 | 164781 | |
| WDR70 | 55100 | ||
| WDR71 | PAAF1; PAAF; Rpn14 | 80227 | |
| WDR72 | AI2A3 | 256764 | Amelogenesis imperfecta |
| WDR73 | HSPC264 | 84942 | |
| WDR74 | 54663 | ||
| WDR75 | NET16; UTP17 | 84128 | |
| WDR76 | CDW14 | 79968 | |
| WDR77 | p44; MEP50; MEP-50; HKMT1069; Nbla10071; p44/Mep50 | 79084 | |
| WDR78 | DIC4 | 79819 | |
| WDR79 | WRAP53; DKCB3; TCAB1 | 55135 | |
| WDR80 | ATG16L; ATG16B | 89849 | |
| WDR81 | CAMRQ2; PPP1R166 | 124997 | cerebellar ataxia, mental retardation, and dysequilibrium syndrome-2 |
| WDR82 | SWD2; MST107; WDR82A; MSTP107; PRO2730; TMEM113; PRO34047 | 80335 | |
| WDR83 | MORG1 | 84292 | |
| WDR84 | PAK1IP1; PIP1; MAK11 | 55003 | |
| WDR85 | DPH7; RRT2; C9orf112 | 92715 | |
| WDR86 | 349136 | ||
| WDR87 | NYD-SP11 | 83889 | |
| WDR88 | PQWD | 126248 | |
| WDR89 | MSTP050; C14orf150 | 112840 | |
| WDR90 | C16orf15; C16orf16; C16orf17; C16orf18; C16orf19 | 197335 | |
| WDR91 | HSPC049 | 29062 | |
| WDR92 | MONAD | 116143 | |
| WDR93 | 56964 | ||
| WDR94 | AMBRA1; DCAF3 | 55626 | |
| WDR96 | CFAP43; C10orf79 | 80217 |
See also
- Beta-propeller
- Tomosyn, a protein two WD40 domains
References
- ↑ PDB: 1erj; "Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast". EMBO J. 19 (12): 3016–27. June 2000. doi:10.1093/emboj/19.12.3016. PMID 10856245.
- ↑ "The ancient regulatory-protein family of WD-repeat proteins". Nature 371 (6495): 297–300. September 1994. doi:10.1038/371297a0. PMID 8090199.
- ↑ 3.0 3.1 "The WD40 repeat: a common architecture for diverse functions". Trends Biochem. Sci. 24 (5): 181–5. May 1999. doi:10.1016/S0968-0004(99)01384-5. PMID 10322433.
- ↑ 4.0 4.1 "WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases". Cell. Mol. Life Sci. 58 (14): 2085–97. December 2001. doi:10.1007/PL00000838. PMID 11814058.
- ↑ "WD40 proteins propel cellular networks.". Trends Biochem. Sci. 35 (10): 565–74. May 2010. doi:10.1016/j.tibs.2010.04.003. PMID 20451393.
- ↑ "Initial sequencing and analysis of the human genome". Nature 409 (6822): 860–921. February 2001. doi:10.1038/35057062. PMID 11237011.
External links
- Eukaryotic Linear Motif resource motif class LIG_APCC_Dbox_1
- Eukaryotic Linear Motif resource motif class LIG_APCC_KENbox_2
- Eukaryotic Linear Motif resource motif class LIG_COP1
- Eukaryotic Linear Motif resource motif class LIG_CRL4_Cdt2_1
- Eukaryotic Linear Motif resource motif class LIG_CRL4_Cdt2_2
- Eukaryotic Linear Motif resource motif class LIG_EH1_1
- Eukaryotic Linear Motif resource motif class LIG_GLEBS_BUB3_1
- Eukaryotic Linear Motif resource motif class LIG_RAPTOR_TOS_1
- Eukaryotic Linear Motif resource motif class LIG_SCF_FBW7_1
- Eukaryotic Linear Motif resource motif class LIG_SCF_FBW7_2
- Eukaryotic Linear Motif resource motif class LIG_SCF-TrCP1_1
- Eukaryotic Linear Motif resource motif class LIG_WRPW_1
- Eukaryotic Linear Motif resource motif class LIG_WRPW_2
- Eukaryotic Linear Motif resource motif class TRG_ER_diArg_1
- Eukaryotic Linear Motif resource motif class TRG_ER_diLys_1
- Eukaryotic Linear Motif resource motif class TRG_Golgi_diPhe_1
- Eukaryotic Linear Motif resource motif class TRG_PTS2

