Biology:RNA polymerase II holoenzyme
RNA polymerase II holoenzyme is a form of eukaryotic RNA polymerase II that is recruited to the promoters of protein-coding genes in living cells.[1][2] It consists of RNA polymerase II, a subset of general transcription factors, and regulatory proteins known as SRB proteins[clarification needed].
RNA polymerase II
RNA polymerase II (also called RNAP II and Pol II) is an enzyme found in eukaryotic cells. It catalyzes the transcription of DNA to synthesize precursors of mRNA and most snRNA and microRNA.[3][4] In humans, RNAP II consists of seventeen protein molecules (gene products encoded by POLR2A-L, where the proteins synthesized from POLR2C, POLR2E, and POLR2F form homodimers).
General transcription factors
General transcription factors (GTFs) or basal transcription factors are protein transcription factors that have been shown to be important in the transcription of class II genes to mRNA templates.[5] Many of them are involved in the formation of a preinitiation complex, which, together with RNA polymerase II, bind to and read the single-stranded DNA gene template.[6] The cluster of RNA polymerase II and various transcription factors is known as a basal transcriptional complex (BTC).
Preinitiation complex
The preinitiation complex (PIC) is a large complex of proteins that is necessary for the transcription of protein-coding genes in eukaryotes and archaea. The PIC helps position RNA polymerase II over gene transcription start sites, denatures the DNA, and positions the DNA in the RNA polymerase II active site for transcription.[5]
The typical PIC is made up of six general transcription factors: TFIIA (GTF2A1, GTF2A2), TFIIB (GTF2B), B-TFIID (BTAF1, TBP), TFIID (BTAF1, BTF3, BTF3L4, EDF1, TAF1-15, 16 total), TFIIE, TFIIF, TFIIH and TFIIJ.
The construction of the polymerase complex takes place on the gene promoter. The TATA box is one well-studied example of a promoter element that occurs in approximately 10% of genes. It is conserved in many (though not all) model eukaryotes and is found in a fraction of the promoters in these organisms. The sequence TATA (or variations) is located at approximately 25 nucleotides upstream of the Transcription Start Point (TSP). In addition, there are also some weakly conserved features including the TFIIB-Recognition Element (BRE), approximately 5 nucleotides upstream (BREu) and 5 nucleotides downstream (BREd) of the TATA box.[7]
Assembly of the PIC
Although the sequence of steps involved in the assembly of the PIC can vary, in general, they follow step 1, binding to the promoter.
- The TATA-binding protein (TBP, a subunit of TFIID), TBPL1, or TBPL2 can bind the promoter or TATA box.[8] Most genes lack a TATA box and use an initiator element (Inr) or downstream core promoter instead. Nevertheless, TBP is always involved and is forced to bind without sequence specificity. TAFs from TFIID can also be involved when the TATA box is absent. A TFIID TAF will bind sequence specifically, and force the TBP to bind non-sequence specifically, bringing the remaining portions of TFIID to the promoter.
- TFIIA interacts with the TBP subunit of TFIID and aids in the binding of TBP to TATA-box containing promoter DNA. Although TFIIA does not recognize DNA itself, its interactions with TBP allow it to stabilize and facilitate formation of the PIC.
- The N-terminal domain of TFIIB brings the DNA into proper position for entry into the active site of RNA polymerase II. TFIIB binds partially sequence specifically, with some preference for BRE. The TFIID-TFIIA-TFIIB (DAB)-promoter complex subsequently recruits RNA polymerase II and TFIIF.[8]
- TFIIF (two subunits, RAP30 and RAP74, showing some similarity to bacterial sigma factors) and Pol II enter the complex together. TFIIF helps to speed up the polymerization process.
- TFIIE joins the growing complex and recruits TFIIH. TFIIE may be involved in DNA melting at the promoter: it contains a zinc ribbon motif that can bind single-stranded DNA.[5] TFIIE helps to open and close the Pol II’s Jaw-like structure, which enables movement down the DNA strand.
- DNA may be wrapped one complete turn around the preinitiation complex and it is TFIIF that helps keep this tight wrapping.[5] In the process, the torsional strain on the DNA may aid in DNA melting at the promoter, forming the transcription bubble.
- TFIIH enters the complex. TFIIH is a large protein complex that contains among others the CDK7/cyclin H kinase complex and a DNA helicase. TFIIH has three functions: It binds specifically to the template strand to ensure that the correct strand of DNA is transcribed and melts or unwinds the DNA (ATP-dependent) to separate the two strands using its helicase activity. It has a kinase activity that phosphorylates the C-terminal domain (CTD) of Pol II at the amino acid serine. This switches the RNA polymerase to start producing RNA.[9] Finally it is essential for Nucleotide Excision Repair (NER) of damaged DNA. TFIIH and TFIIE strongly interact with one another. TFIIE affects TFIIH's catalytic activity. Without TFIIE, TFIIH will not unwind the promoter.
- TFIIH helps create the transcription bubble and may be required for transcription if the DNA template is not already denatured or if it is supercoiled.
- Mediator then encases all the transcription factors and Pol II. It interacts with enhancers, areas very far away (upstream or downstream) that help regulate transcription.[10]
The formation of the preinitiation complex (PIC) is analogous to the mechanism seen in bacterial initiation. In bacteria, the sigma factor recognizes and binds to the promoter sequence. In eukaryotes, the transcription factors perform this role.[9]
Mediator complex
Mediator is a multiprotein complex that functions as a transcriptional coactivator. The Mediator complex is required for the successful transcription of nearly all class II gene promoters in yeast.[11] It works in the same manner in mammals.
The mediator functions as a coactivator and binds to the C-terminal domain (CTD) of RNA polymerase II holoenzyme, acting as a bridge between this enzyme and transcription factors.[12]
C-terminal domain (CTD)
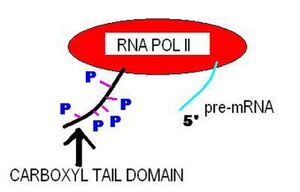
The carboxy-terminal domain (CTD) of RNA polymerase II is that portion of the polymerase that is involved in the initiation of DNA transcription, the capping of the RNA transcript, and attachment to the spliceosome for RNA splicing.[13] The CTD typically consists of up to 52 repeats (in humans) of the sequence Tyr-Ser-Pro-Thr-Ser-Pro-Ser.[14] The carboxy-terminal repeat domain (CTD) is essential for life. Cells containing only RNAPII with none or only up to one-third of its repeats are inviable.[15]
The CTD is an extension appended to the C terminus of RPB1, the largest subunit of RNA polymerase II. It serves as a flexible binding scaffold for numerous nuclear factors, determined by the phosphorylation patterns on the CTD repeats. Each repeat contains an evolutionary conserved and repeated heptapeptide, Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7, which is subjected to reversible phosphorylations during each transcription cycle.[16] This domain is inherently unstructured yet evolutionarily conserved, and in eukaryotes it comprises from 25 to 52 tandem copies of the consensus repeat heptad.[15] As the CTD is frequently not required for general transcription factor (GTF)-mediated initiation and RNA synthesis, it does not form a part of the catalytic essence of RNAPII, but performs other functions.[16]
CTD phosphorylation
RNAPII can exist in two forms: RNAPII0, with a highly phosphorylated CTD, and RNAPIIA, with a nonphosphorylated CTD.[16] Phosphorylation occurs principally on Ser2 and Ser5 of the repeats, although these positions are not equivalent. The phosphorylation state changes as RNAPII progresses through the transcription cycle: The initiating RNAPII is form IIA, and the elongating enzyme is form II0. While RNAPII0 does consist of RNAPs with hyperphosphorylated CTDs, the pattern of phosphorylation on individual CTDs can vary due to differential phosphorylation of Ser2 versus Ser5 residues and/or to differential phosphorylation of repeats along the length of the CTD.[16] The PCTD (phosphoCTD of an RNAPII0) physically links pre-mRNA processing to transcription by tethering processing factors to elongating RNAPII, e.g., 5′-end capping, 3′-end cleavage, and polyadenylation.[16]
Ser5 phosphorylation (Ser5PO4) near the 5′ ends of genes depends principally on the kinase activity of TFIIH (Kin28 in yeast; CDK7 in metazoans).[16] The transcription factor TFIIH is a kinase and will hyperphosphorylate the CTD of RNAP, and in doing so, causes the RNAP complex to move away from the initiation site. Subsequent to the action of TFIIH kinase, Ser2 residues are phosphorylated by CTDK-I in yeast (CDK9 kinase in metazoans). Ctk1 (CDK9) acts in complement to phosphorylation of serine 5 and is, thus, seen in middle to late elongation.
CDK8 and cyclin C (CCNC) are components of the RNA polymerase II holoenzyme that phosphorylate the carboxy-terminal domain (CTD). CDK8 regulates transcription by targeting the CDK7/cyclin H subunits of the general transcription initiation factor IIH (TFIIH), thereby providing a link between the mediator and the basal transcription machinery.[17]
The gene CTDP1 encodes a phosphatase that interacts with the carboxy-terminus of transcription initiation factor TFIIF, a transcription factor that regulates elongation as well as initiation by RNA polymerase II.[18]
Also involved in the phosphorylation and regulation of the RPB1 CTD is cyclin T1 (CCNT1).[19] Cyclin T1 tightly associates and forms a complex with CDK9 kinase, both of which are involved in the phosphorylation and regulation.
- ATP + [DNA-directed RNA polymerase II] <=> ADP + [DNA-directed RNA polymerase II] phosphate : catalyzed by CDK9 EC 2.7.11.23.
TFIIF and FCP1 cooperate for RNAPII recycling. FCP1, the CTD phosphatase, interacts with RNA polymerase II. Transcription is regulated by the state of phosphorylation of a heptapeptide repeat.[20] The nonphosphorylated form, RNAPIIA, is recruited to the initiation complex, whereas the elongating polymerase is found with RNAPII0. RNAPII cycles during transcription. CTD phosphatase activity is regulated by two GTFs (TFIIF and TFIIB). The large subunit of TFIIF (RAP74) stimulates the CTD phosphatase activity, whereas TFIIB inhibits TFIIF-mediated stimulation. Dephosphorylation of the CTD alters the migration of the largest subunit of RNAPII (RPB1).
5' capping
The carboxy-terminal domain is also the binding site of the cap-synthesizing and cap-binding complex. In eukaryotes, after transcription of the 5' end of an RNA transcript, the cap-synthesizing complex on the CTD will remove the gamma-phosphate from the 5'-phosphate and attach a GMP, forming a 5',5'-triphosphate linkage. The synthesizing complex falls off and the cap then binds to the cap-binding complex (CBC), which is bound to the CTD.
The 5'cap of eukaryotic RNA transcripts is important for binding of the mRNA transcript to the ribosome during translation, to the CTD of RNAP, and prevents RNA degradation.
Spliceosome
The carboxy-terminal domain is also the binding site for spliceosome factors that are part of RNA splicing. These allow for the splicing and removal of introns (in the form of a lariat structure) during RNA transcription.
Mutation in the CTD
Major studies in which knockout of particular amino acids was achieved in the CTD have been carried out. The results indicate that RNA polymerase II CTD truncation mutations affect the ability to induce transcription of a subset of genes in vivo, and the lack of response to induction maps to the upstream activating sequences of these genes.
Genome surveillance complex
Several protein members of the BRCA1-associated genome surveillance complex (BASC) associate with RNA polymerase II and play a role in transcription.[21]
The transcription factor TFIIH is involved in transcription initiation and DNA repair. MAT1 (for 'ménage à trois-1') is involved in the assembly of the CAK complex. CAK is a multisubunit protein that includes CDK7, cyclin H (CCNH), and MAT1. CAK is an essential component of the transcription factor TFIIH that is involved in transcription initiation and DNA repair.
The nucleotide excision repair (NER) pathway is a mechanism to repair damage to DNA. ERCC2 is involved in transcription-coupled NER and is an integral member of the basal transcription factor BTF2/TFIIH complex. ERCC3 is an ATP-dependent DNA helicase that functions in NER. It also is a subunit of basal transcription factor 2 (TFIIH) and, thus, functions in class II transcription. XPG (ERCC5) forms a stable complex with TFIIH, which is active in transcription and NER.[22] ERCC6 encodes a DNA-binding protein that is important in transcription-coupled excision repair. ERCC8 interacts with Cockayne syndrome type B (CSB) protein, with p44 (GTF2H2), a subunit of the RNA polymerase II transcription factor IIH, and ERCC6. It is involved in transcription-coupled excision repair.
Higher error ratios in transcription by RNA polymerase II are observed in the presence of Mn2+ compared to Mg2+.[23]
Transcription coactivators
The EDF1 gene encodes a protein that acts as a transcriptional coactivator by interconnecting the general transcription factor TATA element-binding protein (TBP) and gene-specific activators.[24]
TFIID and human mediator coactivator (THRAP3) complexes (mediator complex, plus THRAP3 protein) assemble cooperatively on promoter DNA, from which they become part of the RNAPII holoenzyme.
Transcription initiation
The completed assembly of the holoenzyme with transcription factors and RNA polymerase II bound to the promoter forms the eukaryotic transcription initiation complex. Transcription in the archaea domain is similar to transcription in eukaryotes.[25]
Transcription begins with matching of NTPs to the first and second in the DNA sequence. This, like most of the remainder of transcription, is an energy-dependent process, consuming adenosine triphosphate (ATP) or other NTP.
Promoter clearance
After the first bond is synthesized, the RNA polymerase must clear the promoter. During this time, there is a tendency to release the RNA transcript and produce truncated transcripts. This is called abortive initiation and is common for both eukaryotes and prokaryotes.[26] Abortive initiation continues to occur until the σ factor rearranges, resulting in the transcription elongation complex (which gives a 35 bp-moving footprint). The σ factor is released before 80 nucleotides of mRNA are synthesized.[27] Once the transcript reaches approximately 23 nucleotides, it no longer slips and elongation can occur.
Initiation regulation
Due to the range of genes that Pol II transcribes, this is the polymerase that experiences the most regulation by a range of factors at each stage of transcription. It is also one of the most complex in terms of polymerase cofactors involved.
Initiation is regulated by many mechanisms. These can be separated into two main categories:
- Protein interference.
- Regulation by phosphorylation.
Regulation by protein interference
Protein interference is the process where in some signaling protein interacts, either with the promoter or with some stage of the partially constructed complex, to prevent further construction of the polymerase complex, so preventing initiation. In general, this is a very rapid response and is used for fine level, individual gene control and for 'cascade' processes for a group of genes useful under a specific conditions (for example, DNA repair genes or heat shock genes).
Chromatin structure inhibition is the process wherein the promoter is hidden by chromatin structure. Chromatin structure is controlled by post-translational modification of the histones involved and leads to gross levels of high or low transcription levels. See: chromatin, histone, and nucleosome.
These methods of control can be combined in a modular method, allowing very high specificity in transcription initiation control.
Regulation by phosphorylation
The largest subunit of Pol II (Rpb1) has a domain at its C-terminus called the CTD (C-terminal domain). This is the target of kinases and phosphatases. The phosphorylation of the CTD is an important regulation mechanism, as this allows attraction and rejection of factors that have a function in the transcription process. The CTD can be considered as a platform for transcription factors.
The CTD consists of repetitions of an amino acid motif, YSPTSPS, of which Serines and Threonines can be phosphorylated. The number of these repeats varies; the mammalian protein contains 52, while the yeast protein contains 26. Site-directed-mutagenesis of the yeast protein has found at least 10 repeats are needed for viability. There are many different combinations of phosphorylations possible on these repeats and these can change rapidly during transcription. The regulation of these phosphorylations and the consequences for the association of transcription factors plays a major role in the regulation of transcription.
During the transcription cycle, the CTD of the large subunit of RNAP II is reversibly phosphorylated. RNAP II containing unphosphorylated CTD is recruited to the promoter, whereas the hyperphosphorylated CTD form is involved in active transcription. Phosphorylation occurs at two sites within the heptapeptide repeat, at Serine 5 and Serine 2. Serine 5 phosphorylation is confined to promoter regions and is necessary for the initiation of transcription, whereas Serine 2 phosphorylation is important for mRNA elongation and 3'-end processing.
Elongation
The process of elongation is the synthesis of a copy of the DNA into messenger RNA. RNA Pol II matches complementary RNA nucleotides to the template DNA by Watson-Crick base pairing. These RNA nucleotides are ligated, resulting in a strand of messenger RNA.
Unlike DNA replication, mRNA transcription can involve multiple RNA polymerases on a single DNA template and multiple rounds of transcription (amplification of particular mRNA), so many mRNA molecules can be rapidly produced from a single copy of a gene.
Elongation also involves a proofreading mechanism that can replace incorrectly incorporated bases. In eukaryotes, this may correspond with short pauses during transcription that allow appropriate RNA editing factors to bind. These pauses may be intrinsic to the RNA polymerase or due to chromatin structure.
Elongation regulation
RNA Pol II elongation promoters can be summarised in three classes:
- Drug/sequence-dependent arrest affected factors, e.g., SII (TFIIS) and P-TEFb protein families.
- Chromatin structure oriented factors. Based on histone post translational modifications – phosphorylation, acetylation, methylation and ubiquination.
- See: chromatin, histone, and nucleosome
- RNA Pol II catalysis improving factors. Improve the Vmax or Km of RNA Pol II, so improving the catalytic quality of the polymerase enzyme. E.g. TFIIF, Elongin and ELL families.
- See: Enzyme kinetics, Henri–Michaelis–Menten kinetics, Michaelis constant, and Lineweaver–Burk plot
As for initiation, protein interference, seen as the "drug/sequence-dependent arrest affected factors" and "RNA Pol II catalysis improving factors" provide a very rapid response and is used for fine level individual gene control. Elongation downregulation is also possible, in this case usually by blocking polymerase progress or by deactivating the polymerase.
Chromatin structure-oriented factors are more complex than for initiation control. Often the chromatin-altering factor becomes bound to the polymerase complex, altering the histones as they are encountered and providing a semi-permanent 'memory' of previous promotion and transcription.
Termination
Termination is the process of breaking up the polymerase complex and ending the RNA strand. In eukaryotes using RNA Pol II, this termination is very variable (up to 2000 bases), relying on post transcriptional modification.
Little regulation occurs at termination, although it has been proposed newly transcribed RNA is held in place if proper termination is inhibited, allowing very fast expression of genes given a stimulus. This has not yet been demonstrated in eukaryotes.
Transcription factory
Active RNA Pol II transcription holoenzymes can be clustered in the nucleus, in discrete sites called transcription factories. There are ~8,000 such factories in the nucleoplasm of a HeLa cell, but only 100–300 RNAP II foci per nucleus in erythroid cells, as in many other tissue types.[28] The number of transcription factories in tissues is far more restricted than indicated by previous estimates from cultured cells.[28] As an active transcription unit is usually associated with only one Pol II holoenzyme, a polymerase II factory may contain on average ~8 holoenzymes. Colocalization of transcribed genes has not been observed when using cultured fibroblast-like cells.[29] Differentiated or committed tissue types have a limited number of available transcription sites.[28] Estimates show that erythroid cells express at least 4,000 genes, so many genes are obliged to seek out and share the same factory.[28]
The intranuclear position of many genes is correlated with their activity state. During transcription in vivo, distal active genes are dynamically organized into shared nuclear subcompartments and colocalize to the same transcription factory at high frequencies.[28] Movement into or out of these factories results in activation (On) or abatement (Off) of transcription, rather than by recruiting and assembling a transcription complex.[28] Usually, genes migrate to preassembled factories for transcription.[28]
An expressed gene is preferentially located outside of its chromosome territory, but a closely linked, inactive gene is located inside.[30]
Holoenzyme stability
RNA polymerase II holoenzyme stability determines the number of base pairs that can be transcribed before the holoenzyme loses its ability to transcribe. The length of the CTD is essential for RNA polymerase II stability.[31] RNA polymerase II stability has been shown to be regulated by post-translation proline hydroxylation.[32] The von Hippel–Lindau tumor suppressor protein (pVHL, human GeneID: 7428[33]) complex binds the hyperphosphorylated large subunit of the RNA polymerase II complex, in a proline hydroxylation- and CTD phosphorylation-dependent manner, targeting it for ubiquitination.[32]
See also
- RNA polymerase I
- RNA polymerase III
- Post-transcriptional modification
- Transcription (genetics)
- Eukaryotic transcription
References
- ↑ Koleske, A.J.; Young, R.A. (1994). "An RNA polymerase II holoenzyme responsive to activators". Nature 368 (6470): 466–469. doi:10.1038/368466a0. PMID 8133894. Bibcode: 1994Natur.368..466K.
- ↑ "RNA polymerase II holoenzymes and subcomplexes". J. Biol. Chem. 273 (43): 27757–60. October 1998. doi:10.1074/jbc.273.43.27757. PMID 9774381.
- ↑ Kornberg R (1999). "Eukaryotic transcriptional control". Trends in Cell Biology 9 (12): M46–M49. doi:10.1016/S0962-8924(99)01679-7. PMID 10611681.
- ↑ Sims, R. J. 3rd; Mandal, S. S.; Reinberg, D. (June 2004). "Recent highlights of RNA-polymerase-II-mediated transcription". Current Opinion in Cell Biology 16 (3): 263–271. doi:10.1016/j.ceb.2004.04.004. ISSN 0955-0674. PMID 15145350.
- ↑ 5.0 5.1 5.2 5.3 "Transcription of eukaryotic protein-coding genes". Annu Rev Genet 34: 77–137. 2000. doi:10.1146/annurev.genet.34.1.77. PMID 11092823.
- ↑ "The general transcription factors of RNA polymerase II". Genes Dev. 10 (21): 2657–83. 1996. doi:10.1101/gad.10.21.2657. PMID 8946909.
- ↑ "Polymerase II". http://www.als.lbl.gov/als/science/sci_archive/polymerase2.html.
- ↑ 8.0 8.1 "An RNA Polymerase II Complex Containing All Essential Initiation Factors Binds to the Activation Domain of PAR Leucine Zipper Transcription Factor Thyroid Embryonic Factor". Mol Cell Biol 19 (2): 1242–50. Feb 1999. doi:10.1128/mcb.19.2.1242. PMID 9891058.
- ↑ 9.0 9.1 Pierce, Benjamin C. (2007). Genetics: A Conceptual Approach (3rd ed.). San Francisco: W. H. Freeman. pp. 360–1. ISBN 978-0-7167-7928-5.
- ↑ "Activator-Independent Functions of the Yeast Mediator Sin4 Complex in Preinitiation Complex Formation and Transcription Reinitiation". Mol Cell Biol 23 (1): 349–58. Jan 2003. doi:10.1128/MCB.23.1.349-358.2003. PMID 12482986. PMC 140685. http://biosupport.licor.com./docs/odyssey/pubs/activator_independent_function_of_sin4.pdf.
- ↑ "Yeast mediator and its role in transcriptional regulation". Comptes Rendus Biologies 328 (9): 773–82. 2005. doi:10.1016/j.crvi.2005.03.004. PMID 16168358. https://comptes-rendus.academie-sciences.fr/biologies/articles/10.1016/j.crvi.2005.03.004/.
- ↑ "The yeast Mediator complex and its regulation". Trends Biochem. Sci. 30 (5): 240–4. 2005. doi:10.1016/j.tibs.2005.03.008. PMID 15896741.
- ↑ "Functional studies of the carboxy-terminal repeat domain of Drosophila RNA polymerase II in vivo". Genetics 140 (2): 599–613. June 1995. doi:10.1093/genetics/140.2.599. PMID 7498740.
- ↑ "Recognition of RNA polymerase II carboxy-terminal domain by 3'-RNA-processing factors" (abstract). Nature 430 (6996): 223–6. Jul 2004. doi:10.1038/nature02679. PMID 15241417. Bibcode: 2004Natur.430..223M. http://www.nature.com/nature/journal/v430/n6996/abs/nature02679.htm.
- ↑ 15.0 15.1 Corden JL (1990). "Tails of RNA polymerase II". Trends Biochem. Sci. 15 (10): 383–7. doi:10.1016/0968-0004(90)90236-5. PMID 2251729.
- ↑ 16.0 16.1 16.2 16.3 16.4 16.5 "Phosphorylation and functions of the RNA polymerase II CTD". Genes Dev. 20 (1): 2922–36. Nov 2006. doi:10.1101/gad.1477006. PMID 17079683.
- ↑ "CDK8 cyclin-dependent kinase 8 [Homo sapiens"]. https://www.ncbi.nlm.nih.gov/gene/1024?ordinalpos=1&itool=EntrezSystem2.PEntrez.Gene.Gene_ResultsPanel.Gene_RVDocSum.
- ↑ "CTDP1 CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) phosphatase, subunit 1 [ Homo sapiens "]. https://www.ncbi.nlm.nih.gov/gene/9150?ordinalpos=1&itool=EntrezSystem2.PEntrez.Gene.Gene_ResultsPanel.Gene_RVDocSum.
- ↑ "CCNT1 cyclin T1 [ Homo sapiens ]".
- ↑ "A protein phosphatase functions to recycle RNA polymerase II". Genes Dev. 13 (12): 1540–52. Jun 1999. doi:10.1101/gad.13.12.1540. PMID 10385623.
- ↑ "BRCA1 breast cancer 1, early onset [ Homo sapiens "]. https://www.ncbi.nlm.nih.gov/gene/672?ordinalpos=1&itool=EntrezSystem2.PEntrez.Gene.Gene_ResultsPanel.Gene_RVDocSum.
- ↑ "Interactions involving the human RNA polymerase II transcription/nucleotide excision repair complex TFIIH, the nucleotide excision repair protein XPG, and Cockayne syndrome group B (CSB) protein". Biochemistry 35 (7): 2157–67. 1996. doi:10.1021/bi9524124. PMID 8652557.
- ↑ "DNA-Directed RNA polymerase". DNA-directed RNA polymerase in: Springer Handbook of Enzymes. 38. Berlin: Springer. 2007. pp. 103–17. doi:10.1007/978-3-540-71526-9_11. ISBN 978-3-540-71525-2.
- ↑ "EDF1 endothelial differentiation-related factor 1 [ Homo sapiens "]. https://www.ncbi.nlm.nih.gov/gene/8721?ordinalpos=1&itool=EntrezSystem2.PEntrez.Gene.Gene_ResultsPanel.Gene_RVDocSum.
- ↑ "Activation of archaeal transcription by recruitment of the TATA-binding protein". Proc. Natl. Acad. Sci. U.S.A. 100 (9): 5097–102. 2003. doi:10.1073/pnas.0837150100. PMID 12692306. Bibcode: 2003PNAS..100.5097O.
- ↑ Goldman, S.; Ebright, R.; Nickels, B. (May 2009). "Direct detection of abortive RNA transcripts in vivo". Science 324 (5929): 927–928. doi:10.1126/science.1169237. PMID 19443781. Bibcode: 2009Sci...324..927G.
- ↑ Dvir, A. (Sep 2002). "Promoter escape by RNA polymerase II". Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1577 (2): 208–223. doi:10.1016/s0167-4781(02)00453-0. ISSN 0006-3002. PMID 12213653.
- ↑ 28.0 28.1 28.2 28.3 28.4 28.5 28.6 "Active genes dynamically colocalize to shared sites of ongoing transcription". Nature Genetics 36 (10): 1065–71. Oct 2001. doi:10.1038/ng1423. PMID 15361872.
- ↑ "Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods". J. Cell Biol. 162 (6): 981–90. 2003. doi:10.1083/jcb.200303131. PMID 12975345.
- ↑ "Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription". Genes Dev. 18 (10): 1119–30. 2004. doi:10.1101/gad.292104. PMID 15155579.
- ↑ "An Unusual Recent Expansion of the C-Terminal Domain of RNA Polymerase II in Primate Malaria Parasites Features a Motif Otherwise Found Only in Mammalian Polymerases". J Mol Evol 68 (6): 706–14. Jun 2009. doi:10.1007/s00239-009-9245-2. PMID 19449052. Bibcode: 2009JMolE..68..706K.
- ↑ 32.0 32.1 "Von Hippel-Lindau protein binds hyperphosphorylated large subunit of RNA polymerase II through a proline hydroxylation motif and targets it for ubiquitination". Proc. Natl. Acad. Sci. U.S.A. 100 (5): 2706–11. Mar 2003. doi:10.1073/pnas.0436037100. PMID 12604794. Bibcode: 2003PNAS..100.2706K.
- ↑ "VHL von Hippel-Lindau tumor suppressor [ Homo sapiens "]. https://www.ncbi.nlm.nih.gov/gene/7428?ordinalpos=1&itool=EntrezSystem2.PEntrez.Gene.Gene_ResultsPanel.Gene_RVDocSum.
- "A kinase–cyclin pair in the RNA polymerase II holoenzyme". Nature 374 (6518): 193–6. 2002. doi:10.1038/374193a0. PMID 7877695.
- RNA Polymerase: Components of the Transcription Initiation Machinery
- "Factors associated with the mammalian RNA polymerase II holoenzyme". Nucleic Acids Res. 26 (3): 847–53. February 1998. doi:10.1093/nar/26.3.847. PMID 9443979.
- "General requirement for RNA polymerase II holoenzymes in vivo". Proc. Natl. Acad. Sci. U.S.A. 92 (10): 4587–90. May 1995. doi:10.1073/pnas.92.10.4587. PMID 7753848. Bibcode: 1995PNAS...92.4587T.
External links
- More information at Berkeley National Lab
- RNA+Polymerase+II at the US National Library of Medicine Medical Subject Headings (MeSH)
 |

