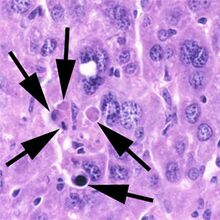
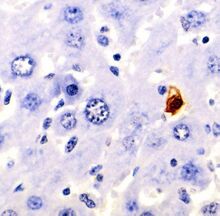
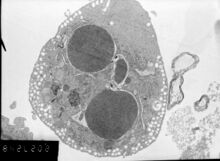
Biology:Apoptosis
| Apoptosis | |
|---|---|
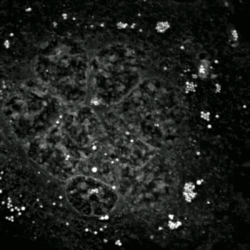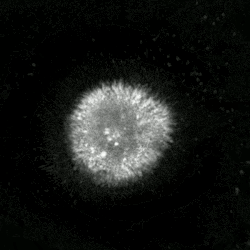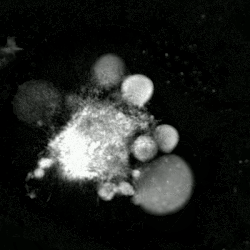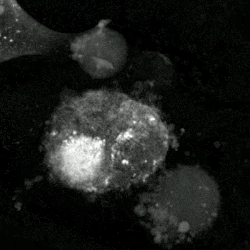
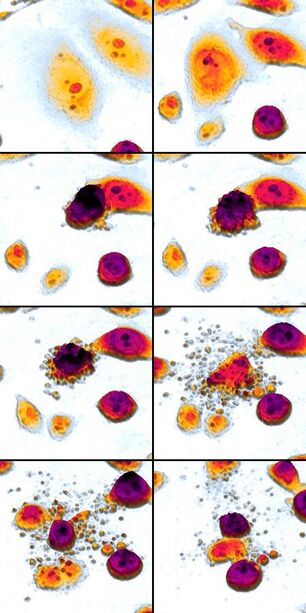 An etoposide-treated DU145 prostate cancer cell exploding into a cascade of apoptotic bodies. The sub images were extracted from a 61-hour time-lapse microscopy video, created using quantitative phase-contrast microscopy. The optical thickness is color-coded. With increasing thickness, color changes from gray to yellow, red, purple and finally black. See the video at The Cell: An Image Library | |
| Anatomical terminology |
Apoptosis (from Ancient Greek:) is a form of programmed cell death that occurs in multicellular organisms and in some eukaryotic, single-celled microorganisms such as yeast.[1] Biochemical events lead to characteristic cell changes (morphology) and death.[2] These changes include blebbing, cell shrinkage, nuclear fragmentation, chromatin condensation, DNA fragmentation, and mRNA decay. The average adult human loses between 50 and 70 billion cells each day due to apoptosis.[lower-alpha 1] For an average human child between eight and fourteen years old, each day the approximate loss is 20 to 30 billion cells.[4]
In contrast to necrosis, which is a form of traumatic cell death that results from acute cellular injury, apoptosis is a highly regulated and controlled process that confers advantages during an organism's life cycle. For example, the separation of fingers and toes in a developing human embryo occurs because cells between the digits undergo apoptosis. Unlike necrosis, apoptosis produces cell fragments called apoptotic bodies that phagocytes are able to engulf and remove before the contents of the cell can spill out onto surrounding cells and cause damage to them.[5]
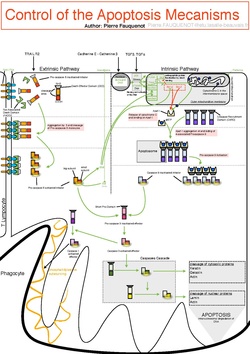
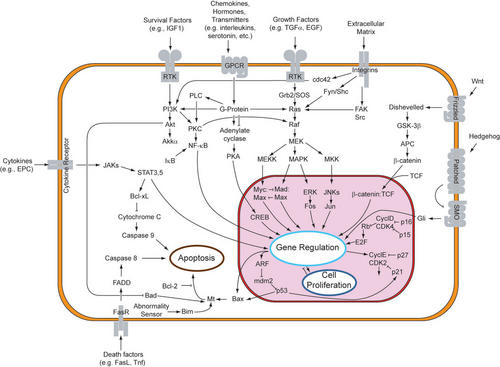
Because apoptosis cannot stop once it has begun, it is a highly regulated process. Apoptosis can be initiated through one of two pathways. In the intrinsic pathway the cell kills itself because it senses cell stress, while in the extrinsic pathway the cell kills itself because of signals from other cells. Weak external signals may also activate the intrinsic pathway of apoptosis.[6] Both pathways induce cell death by activating caspases, which are proteases, or enzymes that degrade proteins. The two pathways both activate initiator caspases, which then activate executioner caspases, which then kill the cell by degrading proteins indiscriminately.
In addition to its importance as a biological phenomenon, defective apoptotic processes have been implicated in a wide variety of diseases. Excessive apoptosis causes atrophy, whereas an insufficient amount results in uncontrolled cell proliferation, such as cancer. Some factors like Fas receptors and caspases promote apoptosis, while some members of the Bcl-2 family of proteins inhibit apoptosis.[7]
Discovery and etymology
German scientist Carl Vogt was first to describe the principle of apoptosis in 1842. In 1885, anatomist Walther Flemming delivered a more precise description of the process of programmed cell death. However, it was not until 1965 that the topic was resurrected. While studying tissues using electron microscopy, John Kerr at the University of Queensland was able to distinguish apoptosis from traumatic cell death.[8] Following the publication of a paper describing the phenomenon, Kerr was invited to join Alastair Currie, as well as Andrew Wyllie, who was Currie's graduate student,[9] at University of Aberdeen. In 1972, the trio published a seminal article in the British Journal of Cancer.[10] Kerr had initially used the term programmed cell necrosis, but in the article, the process of natural cell death was called apoptosis. Kerr, Wyllie and Currie credited James Cormack, a professor of Greek language at University of Aberdeen, with suggesting the term apoptosis. Kerr received the Paul Ehrlich and Ludwig Darmstaedter Prize on March 14, 2000, for his description of apoptosis. He shared the prize with Boston biologist H. Robert Horvitz.[11]
For many years, neither "apoptosis" nor "programmed cell death" was a highly cited term. Two discoveries brought cell death from obscurity to a major field of research: identification of the first component of the cell death control and effector mechanisms, and linkage of abnormalities in cell death to human disease, in particular cancer. This occurred in 1988 when it was shown that BCL2, the gene responsible for follicular lymphoma, encoded a protein that inhibited cell death.[12]
The 2002 Nobel Prize in Medicine was awarded to Sydney Brenner, H. Robert Horvitz and John Sulston for their work identifying genes that control apoptosis. The genes were identified by studies in the nematode C. elegans and homologues of these genes function in humans to regulate apoptosis.
In Greek, apoptosis translates to the "falling off" of leaves from a tree.[13] Cormack, professor of Greek language, reintroduced the term for medical use as it had a medical meaning for the Greeks over two thousand years before. Hippocrates used the term to mean "the falling off of the bones". Galen extended its meaning to "the dropping of the scabs". Cormack was no doubt aware of this usage when he suggested the name. Debate continues over the correct pronunciation, with opinion divided between a pronunciation with the second p silent (/æpəˈtoʊsɪs/ ap-ə-TOH-sis[14][15]) and the second p pronounced (/eɪpəpˈtoʊsɪs/).[14][16] In English, the p of the Greek -pt- consonant cluster is typically silent at the beginning of a word (e.g. pterodactyl, Ptolemy), but articulated when used in combining forms preceded by a vowel, as in helicopter or the orders of insects: diptera, lepidoptera, etc.
In the original Kerr, Wyllie & Currie paper,[10] there is a footnote regarding the pronunciation:
We are most grateful to Professor James Cormack of the Department of Greek, University of Aberdeen, for suggesting this term. The word "apoptosis" (ἀπόπτωσις) is used in Greek to describe the "dropping off" or "falling off" of petals from flowers, or leaves from trees. To show the derivation clearly, we propose that the stress should be on the penultimate syllable, the second half of the word being pronounced like "ptosis" (with the "p" silent), which comes from the same root "to fall", and is already used to describe the drooping of the upper eyelid.
Activation mechanisms
The initiation of apoptosis is tightly regulated by activation mechanisms, because once apoptosis has begun, it inevitably leads to the death of the cell.[17][2] The two best-understood activation mechanisms are the intrinsic pathway (also called the mitochondrial pathway) and the extrinsic pathway.[18] The intrinsic pathway is activated by intracellular signals generated when cells are stressed and depends on the release of proteins from the intermembrane space of mitochondria.[19] The extrinsic pathway is activated by extracellular ligands binding to cell-surface death receptors, which leads to the formation of the death-inducing signaling complex (DISC).[20]
A cell initiates intracellular apoptotic signaling in response to a stress,[21] which may bring about cell suicide. The binding of nuclear receptors by glucocorticoids,[22] heat,[22] radiation,[22] nutrient deprivation,[22] viral infection,[22] hypoxia,[22] increased intracellular concentration of free fatty acids[23] and increased intracellular calcium concentration,[24][25] for example, by damage to the membrane, can all trigger the release of intracellular apoptotic signals by a damaged cell. A number of cellular components, such as poly ADP ribose polymerase, may also help regulate apoptosis.[26] Single cell fluctuations have been observed in experimental studies of stress induced apoptosis.[27][28]
Before the actual process of cell death is precipitated by enzymes, apoptotic signals must cause regulatory proteins to initiate the apoptosis pathway. This step allows those signals to cause cell death, or the process to be stopped, should the cell no longer need to die. Several proteins are involved, but two main methods of regulation have been identified: the targeting of mitochondria functionality,[29] or directly transducing the signal via adaptor proteins to the apoptotic mechanisms. An extrinsic pathway for initiation identified in several toxin studies is an increase in calcium concentration within a cell caused by drug activity, which also can cause apoptosis via a calcium binding protease calpain.
Intrinsic pathway
The intrinsic pathway is also known as the mitochondrial pathway. Mitochondria are essential to multicellular life. Without them, a cell ceases to respire aerobically and quickly dies. This fact forms the basis for some apoptotic pathways. Apoptotic proteins that target mitochondria affect them in different ways. They may cause mitochondrial swelling through the formation of membrane pores, or they may increase the permeability of the mitochondrial membrane and cause apoptotic effectors to leak out.[22][30] There is also a growing body of evidence indicating that nitric oxide is able to induce apoptosis by helping to dissipate the membrane potential of mitochondria and therefore make it more permeable.[31] Nitric oxide has been implicated in initiating and inhibiting apoptosis through its possible action as a signal molecule of subsequent pathways that activate apoptosis.[32]
During apoptosis, cytochrome c is released from mitochondria through the actions of the proteins Bax and Bak. The mechanism of this release is enigmatic, but appears to stem from a multitude of Bax/Bak homo- and hetero-dimers of Bax/Bak inserted into the outer membrane.[33] Once cytochrome c is released it binds with Apoptotic protease activating factor – 1 (Apaf-1) and ATP, which then bind to pro-caspase-9 to create a protein complex known as an apoptosome. The apoptosome cleaves the pro-caspase to its active form of caspase-9, which in turn cleaves and activates pro-caspase into the effector caspase-3.
Mitochondria also release proteins known as SMACs (second mitochondria-derived activator of caspases) into the cell's cytosol following the increase in permeability of the mitochondria membranes. SMAC binds to proteins that inhibit apoptosis (IAPs) thereby deactivating them, and preventing the IAPs from arresting the process and therefore allowing apoptosis to proceed. IAP also normally suppresses the activity of a group of cysteine proteases called caspases,[34] which carry out the degradation of the cell. Therefore, the actual degradation enzymes can be seen to be indirectly regulated by mitochondrial permeability.
Extrinsic pathway
Two theories of the direct initiation of apoptotic mechanisms in mammals have been suggested: the TNF-induced (tumor necrosis factor) model and the Fas-Fas ligand-mediated model, both involving receptors of the TNF receptor (TNFR) family[35] coupled to extrinsic signals.
TNF pathway
TNF-alpha is a cytokine produced mainly by activated macrophages, and is the major extrinsic mediator of apoptosis. Most cells in the human body have two receptors for TNF-alpha: TNFR1 and TNFR2. The binding of TNF-alpha to TNFR1 has been shown to initiate the pathway that leads to caspase activation via the intermediate membrane proteins TNF receptor-associated death domain (TRADD) and Fas-associated death domain protein (FADD). cIAP1/2 can inhibit TNF-α signaling by binding to TRAF2. FLIP inhibits the activation of caspase-8.[36] Binding of this receptor can also indirectly lead to the activation of transcription factors involved in cell survival and inflammatory responses.[37] However, signalling through TNFR1 might also induce apoptosis in a caspase-independent manner.[38] The link between TNF-alpha and apoptosis shows why an abnormal production of TNF-alpha plays a fundamental role in several human diseases, especially in autoimmune diseases. The TNF-alpha receptor superfamily also includes death receptors (DRs), such as DR4 and DR5. These receptors bind to the protein TRAIL and mediate apoptosis. Apoptosis is known to be one of the primary mechanisms of targeted cancer therapy.[39] Luminescent iridium complex-peptide hybrids (IPHs) have recently been designed, which mimic TRAIL and bind to death receptors on cancer cells, thereby inducing their apoptosis.[40]
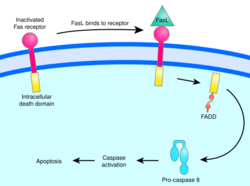
Fas pathway
The fas receptor (First apoptosis signal) – (also known as Apo-1 or CD95) is a transmembrane protein of the TNF family which binds the Fas ligand (FasL).[35] The interaction between Fas and FasL results in the formation of the death-inducing signaling complex (DISC), which contains the FADD, caspase-8 and caspase-10. In some types of cells (type I), processed caspase-8 directly activates other members of the caspase family, and triggers the execution of apoptosis of the cell. In other types of cells (type II), the Fas-DISC starts a feedback loop that spirals into increasing release of proapoptotic factors from mitochondria and the amplified activation of caspase-8.[41]
Common components
Following TNF-R1 and Fas activation in mammalian cells[citation needed] a balance between proapoptotic (BAX,[42] BID, BAK, or BAD) and anti-apoptotic (Bcl-Xl and Bcl-2) members of the Bcl-2 family are established. This balance is the proportion of proapoptotic homodimers that form in the outer-membrane of the mitochondrion. The proapoptotic homodimers are required to make the mitochondrial membrane permeable for the release of caspase activators such as cytochrome c and SMAC. Control of proapoptotic proteins under normal cell conditions of nonapoptotic cells is incompletely understood, but in general, Bax or Bak are activated by the activation of BH3-only proteins, part of the Bcl-2 family.[43]
Caspases
Caspases play the central role in the transduction of ER apoptotic signals. Caspases are proteins that are highly conserved, cysteine-dependent aspartate-specific proteases. There are two types of caspases: initiator caspases (caspases 2, 8, 9, 10, 11, and 12) and effector caspases (caspases 3, 6, and 7). The activation of initiator caspases requires binding to specific oligomeric activator protein. Effector caspases are then activated by these active initiator caspases through proteolytic cleavage. The active effector caspases then proteolytically degrade a host of intracellular proteins to carry out the cell death program.
Caspase-independent apoptotic pathway
There also exists a caspase-independent apoptotic pathway that is mediated by AIF (apoptosis-inducing factor).[44]
Apoptosis model in amphibians
The frog Xenopus laevis serves as an ideal model system for the study of the mechanisms of apoptosis. In fact, iodine and thyroxine also stimulate the spectacular apoptosis of the cells of the larval gills, tail and fins in amphibian's metamorphosis, and stimulate the evolution of their nervous system transforming the aquatic, vegetarian tadpole into the terrestrial, carnivorous frog.[45][46][47][48]
Negative regulators of apoptosis
Negative regulation of apoptosis inhibits cell death signaling pathways, helping tumors to evade cell death and developing drug resistance. The ratio between anti-apoptotic (Bcl-2) and pro-apoptotic (Bax) proteins determines whether a cell lives or dies.[49][50] Many families of proteins act as negative regulators categorized into either antiapoptotic factors, such as IAPs and Bcl-2 proteins or prosurvival factors like cFLIP, BNIP3, FADD, Akt, and NF-κB.[51]
Proteolytic caspase cascade: Killing the cell
Many pathways and signals lead to apoptosis, but these converge on a single mechanism that actually causes the death of the cell. After a cell receives stimulus, it undergoes organized degradation of cellular organelles by activated proteolytic caspases. In addition to the destruction of cellular organelles, mRNA is rapidly and globally degraded by a mechanism that is not yet fully characterized.[52] mRNA decay is triggered very early in apoptosis.
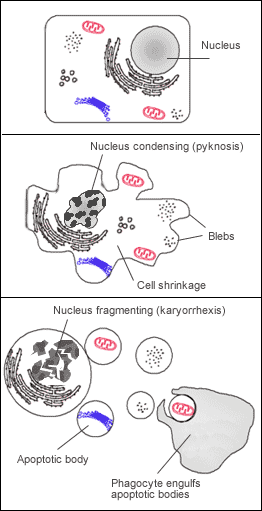
A cell undergoing apoptosis shows a series of characteristic morphological changes. Early alterations include:
- Cell shrinkage and rounding occur because of the retraction lamellipodia and the breakdown of the proteinaceous cytoskeleton by caspases.[53]
- The cytoplasm appears dense, and the organelles appear tightly packed.
- Chromatin undergoes condensation into compact patches against the nuclear envelope (also known as the perinuclear envelope) in a process known as pyknosis, a hallmark of apoptosis.[54][55]
- The nuclear envelope becomes discontinuous and the DNA inside it is fragmented in a process referred to as karyorrhexis. The nucleus breaks into several discrete chromatin bodies or nucleosomal units due to the degradation of DNA.[56]
Apoptosis progresses quickly and its products are quickly removed, making it difficult to detect or visualize on classical histology sections. During karyorrhexis, endonuclease activation leaves short DNA fragments, regularly spaced in size. These give a characteristic "laddered" appearance on agar gel after electrophoresis.[57] Tests for DNA laddering differentiate apoptosis from ischemic or toxic cell death.[58]
Apoptotic cell disassembly
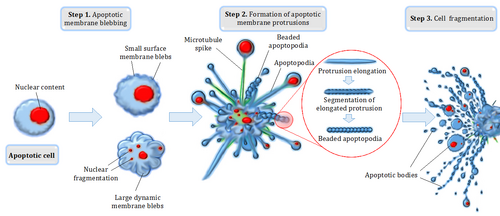
Before the apoptotic cell is disposed of, there is a process of disassembly. There are three recognized steps in apoptotic cell disassembly:[60]
- Membrane blebbing: The cell membrane shows irregular buds known as blebs. Initially these are smaller surface blebs. Later these can grow into larger so-called dynamic membrane blebs.[60] An important regulator of apoptotic cell membrane blebbing is ROCK1 (rho associated coiled-coil-containing protein kinase 1).[61][62]
- Formation of membrane protrusions: Some cell types, under specific conditions, may develop different types of long, thin extensions of the cell membrane called membrane protrusions. Three types have been described: microtubule spikes, apoptopodia (feet of death), and beaded apoptopodia (the latter having a beads-on-a-string appearance).[63][64][65] Pannexin 1 is an important component of membrane channels involved in the formation of apoptopodia and beaded apoptopodia.[64]
- Fragmentation: The cell breaks apart into multiple vesicles called apoptotic bodies, which undergo phagocytosis. The plasma membrane protrusions may help bring apoptotic bodies closer to phagocytes.
Removal of dead cells
The removal of dead cells by neighboring phagocytic cells has been termed efferocytosis.[66] Dying cells that undergo the final stages of apoptosis display phagocytotic molecules, such as phosphatidylserine, on their cell surface.[67] Phosphatidylserine is normally found on the inner leaflet surface of the plasma membrane, but is redistributed during apoptosis to the extracellular surface by a protein known as scramblase.[68] These molecules mark the cell for phagocytosis by cells possessing the appropriate receptors, such as macrophages.[69] The removal of dying cells by phagocytes occurs in an orderly manner without eliciting an inflammatory response.[70] During apoptosis cellular RNA and DNA are separated from each other and sorted to different apoptotic bodies; separation of RNA is initiated as nucleolar segregation.[71]
Pathway knock-outs
Many knock-outs have been made in the apoptosis pathways to test the function of each of the proteins. Several caspases, in addition to APAF1 and FADD, have been mutated to determine the new phenotype. In order to create a tumor necrosis factor (TNF) knockout, an exon containing the nucleotides 3704–5364 was removed from the gene.[72] This exon encodes a portion of the mature TNF domain, as well as the leader sequence, which is a highly conserved region necessary for proper intracellular processing. TNF-/- mice develop normally and have no gross structural or morphological abnormalities. However, upon immunization with SRBC (sheep red blood cells), these mice demonstrated a deficiency in the maturation of an antibody response; they were able to generate normal levels of IgM, but could not develop specific IgG levels.[73] Apaf-1 is the protein that turns on caspase 9 by cleavage to begin the caspase cascade that leads to apoptosis.[74] Since a -/- mutation in the APAF-1 gene is embryonic lethal, a gene trap strategy was used in order to generate an APAF-1 -/- mouse. This assay is used to disrupt gene function by creating an intragenic gene fusion. When an APAF-1 gene trap is introduced into cells, many morphological changes occur, such as spina bifida, the persistence of interdigital webs, and open brain.[75] In addition, after embryonic day 12.5, the brain of the embryos showed several structural changes. APAF-1 cells are protected from apoptosis stimuli such as irradiation. A BAX-1 knock-out mouse exhibits normal forebrain formation and a decreased programmed cell death in some neuronal populations and in the spinal cord, leading to an increase in motor neurons.[76]
The caspase proteins are integral parts of the apoptosis pathway, so it follows that knock-outs made have varying damaging results. A caspase 9 knock-out leads to a severe brain malformation [citation needed]. A caspase 8 knock-out leads to cardiac failure and thus embryonic lethality [citation needed]. However, with the use of cre-lox technology, a caspase 8 knock-out has been created that exhibits an increase in peripheral T cells, an impaired T cell response, and a defect in neural tube closure [citation needed]. These mice were found to be resistant to apoptosis mediated by CD95, TNFR, etc. but not resistant to apoptosis caused by UV irradiation, chemotherapeutic drugs, and other stimuli. Finally, a caspase 3 knock-out was characterized by ectopic cell masses in the brain and abnormal apoptotic features such as membrane blebbing or nuclear fragmentation [citation needed]. A remarkable feature of these KO mice is that they have a very restricted phenotype: Casp3, 9, APAF-1 KO mice have deformations of neural tissue and FADD and Casp 8 KO showed defective heart development, however, in both types of KO other organs developed normally and some cell types were still sensitive to apoptotic stimuli suggesting that unknown proapoptotic pathways exist.
Methods for distinguishing apoptotic from necrotic cells
Label-free live cell imaging, time-lapse microscopy, flow fluorocytometry, and transmission electron microscopy can be used to compare apoptotic and necrotic cells. There are also various biochemical techniques for analysis of cell surface markers (phosphatidylserine exposure versus cell permeability by flow cytometry), cellular markers such as DNA fragmentation[77] (flow cytometry),[78] caspase activation, Bid cleavage, and cytochrome c release (Western blotting). Supernatant screening for caspases, HMGB1, and cytokeratin 18 release can identify primary from secondary necrotic cells. However, no distinct surface or biochemical markers of necrotic cell death have been identified yet, and only negative markers are available. These include absence of apoptotic markers (caspase activation, cytochrome c release, and oligonucleosomal DNA fragmentation) and differential kinetics of cell death markers (phosphatidylserine exposure and cell membrane permeabilization). A selection of techniques that can be used to distinguish apoptosis from necroptotic cells could be found in these references.[79][80][81][82]
Implication in disease
Defective pathways
The many different types of apoptotic pathways contain a multitude of different biochemical components, many of them not yet understood.[83] As a pathway is more or less sequential in nature, removing or modifying one component leads to an effect in another. In a living organism, this can have disastrous effects, often in the form of disease or disorder. A discussion of every disease caused by modification of the various apoptotic pathways would be impractical, but the concept overlying each one is the same: The normal functioning of the pathway has been disrupted in such a way as to impair the ability of the cell to undergo normal apoptosis. This results in a cell that lives past its "use-by date" and is able to replicate and pass on any faulty machinery to its progeny, increasing the likelihood of the cell's becoming cancerous or diseased.
A recently described example of this concept in action can be seen in the development of a lung cancer called NCI-H460.[84] The X-linked inhibitor of apoptosis protein (XIAP) is overexpressed in cells of the H460 cell line. XIAPs bind to the processed form of caspase-9 and suppress the activity of apoptotic activator cytochrome c, therefore overexpression leads to a decrease in the number of proapoptotic agonists. As a consequence, the balance of anti-apoptotic and proapoptotic effectors is upset in favour of the former, and the damaged cells continue to replicate despite being directed to die. Defects in regulation of apoptosis in cancer cells occur often at the level of control of transcription factors. As a particular example, defects in molecules that control transcription factor NF-κB in cancer change the mode of transcriptional regulation and the response to apoptotic signals, to curtail dependence on the tissue that the cell belongs. This degree of independence from external survival signals, can enable cancer metastasis.[85]
Dysregulation of p53
The tumor-suppressor protein p53 accumulates when DNA is damaged due to a chain of biochemical factors. Part of this pathway includes alpha-interferon and beta-interferon, which induce transcription of the p53 gene, resulting in the increase of p53 protein level and enhancement of cancer cell-apoptosis.[86] p53 prevents the cell from replicating by stopping the cell cycle at G1, or interphase, to give the cell time to repair; however, it will induce apoptosis if damage is extensive and repair efforts fail.[87] Any disruption to the regulation of the p53 or interferon genes will result in impaired apoptosis and the possible formation of tumors.
Inhibition
Inhibition of apoptosis can result in a number of cancers, inflammatory diseases, and viral infections. It was originally believed that the associated accumulation of cells was due to an increase in cellular proliferation, but it is now known that it is also due to a decrease in cell death. The most common of these diseases is cancer, the disease of excessive cellular proliferation, which is often characterized by an overexpression of IAP family members. As a result, the malignant cells experience an abnormal response to apoptosis induction: Cycle-regulating genes (such as p53, ras or c-myc) are mutated or inactivated in diseased cells, and further genes (such as bcl-2) also modify their expression in tumors. Some apoptotic factors are vital during mitochondrial respiration e.g. cytochrome C.[88] Pathological inactivation of apoptosis in cancer cells is correlated with frequent respiratory metabolic shifts toward glycolysis (an observation known as the "Warburg hypothesis".[89]
HeLa cell
Apoptosis in HeLa[lower-alpha 2] cells is inhibited by proteins produced by the cell; these inhibitory proteins target retinoblastoma tumor-suppressing proteins.[90] These tumor-suppressing proteins regulate the cell cycle, but are rendered inactive when bound to an inhibitory protein.[90] HPV E6 and E7 are inhibitory proteins expressed by the human papillomavirus, HPV being responsible for the formation of the cervical tumor from which HeLa cells are derived.[91] HPV E6 causes p53, which regulates the cell cycle, to become inactive.[92] HPV E7 binds to retinoblastoma tumor suppressing proteins and limits its ability to control cell division.[92] These two inhibitory proteins are partially responsible for HeLa cells' immortality by inhibiting apoptosis to occur.[93]
Treatments
The main method of treatment for potential death from signaling-related diseases involves either increasing or decreasing the susceptibility of apoptosis in diseased cells, depending on whether the disease is caused by either the inhibition of or excess apoptosis. For instance, treatments aim to restore apoptosis to treat diseases with deficient cell death and to increase the apoptotic threshold to treat diseases involved with excessive cell death. To stimulate apoptosis, one can increase the number of death receptor ligands (such as TNF or TRAIL), antagonize the anti-apoptotic Bcl-2 pathway, or introduce Smac mimetics to inhibit the inhibitor (IAPs).[49] The addition of agents such as Herceptin, Iressa, or Gleevec works to stop cells from cycling and causes apoptosis activation by blocking growth and survival signaling further upstream. Finally, adding p53-MDM2 complexes displaces p53 and activates the p53 pathway, leading to cell cycle arrest and apoptosis. Many different methods can be used either to stimulate or to inhibit apoptosis in various places along the death signaling pathway.[94]
Apoptosis is a multi-step, multi-pathway cell-death programme that is inherent in every cell of the body. In cancer, the apoptosis cell-division ratio is altered. Cancer treatment by chemotherapy and irradiation kills target cells primarily by inducing apoptosis.
Hyperactive apoptosis
On the other hand, loss of control of cell death (resulting in excess apoptosis) can lead to neurodegenerative diseases, hematologic diseases, and tissue damage. Neurons that rely on mitochondrial respiration undergo apoptosis in neurodegenerative diseases such as Alzheimer's[95] and Parkinson's.[96] (an observation known as the "Inverse Warburg hypothesis"[88][97]). Moreover, there is an inverse epidemiological comorbidity between neurodegenerative diseases and cancer.[98] The progression of HIV is directly linked to excess, unregulated apoptosis. In a healthy individual, the number of CD4+ lymphocytes is in balance with the cells generated by the bone marrow; however, in HIV-positive patients, this balance is lost due to an inability of the bone marrow to regenerate CD4+ cells. In the case of HIV, CD4+ lymphocytes die at an accelerated rate through uncontrolled apoptosis, when stimulated. At the molecular level, hyperactive apoptosis can be caused by defects in signaling pathways that regulate the Bcl-2 family proteins. Increased expression of apoptotic proteins such as BIM, or their decreased proteolysis, leads to cell death and can cause a number of pathologies, depending on the cells where excessive activity of BIM occurs. Cancer cells can escape apoptosis through mechanisms that suppress BIM expression or by increased proteolysis of BIM.[citation needed]
Treatments
Treatments aiming to inhibit works to block specific caspases. Finally, the Akt protein kinase promotes cell survival through two pathways. Akt phosphorylates and inhibits Bad (a Bcl-2 family member), causing Bad to interact with the 14-3-3 scaffold, resulting in Bcl dissociation and thus cell survival. Akt also activates IKKα, which leads to NF-κB activation and cell survival. Active NF-κB induces the expression of anti-apoptotic genes such as Bcl-2, resulting in inhibition of apoptosis. NF-κB has been found to play both an antiapoptotic role and a proapoptotic role depending on the stimuli utilized and the cell type.[99]
HIV progression
The progression of the human immunodeficiency virus infection into AIDS is due primarily to the depletion of CD4+ T-helper lymphocytes in a manner that is too rapid for the body's bone marrow to replenish the cells, leading to a compromised immune system. One of the mechanisms by which T-helper cells are depleted is apoptosis, which results from a series of biochemical pathways:[100]
- HIV enzymes deactivate anti-apoptotic Bcl-2. This does not directly cause cell death but primes the cell for apoptosis should the appropriate signal be received. In parallel, these enzymes activate proapoptotic procaspase-8, which does directly activate the mitochondrial events of apoptosis.
- HIV may increase the level of cellular proteins that prompt Fas-mediated apoptosis.
- HIV proteins decrease the amount of CD4 glycoprotein marker present on the cell membrane.
- Released viral particles and proteins present in extracellular fluid are able to induce apoptosis in nearby "bystander" T helper cells.
- HIV decreases the production of molecules involved in marking the cell for apoptosis, giving the virus time to replicate and continue releasing apoptotic agents and virions into the surrounding tissue.
- The infected CD4+ cell may also receive the death signal from a cytotoxic T cell.
Cells may also die as direct consequences of viral infections. HIV-1 expression induces tubular cell G2/M arrest and apoptosis.[101] The progression from HIV to AIDS is not immediate or even necessarily rapid; HIV's cytotoxic activity toward CD4+ lymphocytes is classified as AIDS once a given patient's CD4+ cell count falls below 200.[102]
Researchers from Kumamoto University in Japan have developed a new method to eradicate HIV in viral reservoir cells, named "Lock-in and apoptosis." Using the synthesized compound Heptanoylphosphatidyl L-Inositol Pentakisphophate (or L-Hippo) to bind strongly to the HIV protein PR55Gag, they were able to suppress viral budding. By suppressing viral budding, the researchers were able to trap the HIV virus in the cell and allow for the cell to undergo apoptosis (natural cell death). Associate Professor Mikako Fujita has stated that the approach is not yet available to HIV patients because the research team has to conduct further research on combining the drug therapy that currently exists with this "Lock-in and apoptosis" approach to lead to complete recovery from HIV.[103]
Viral infection
Viral induction of apoptosis occurs when one or several cells of a living organism are infected with a virus, leading to cell death. Cell death in organisms is necessary for the normal development of cells and the cell cycle maturation.[104] It is also important in maintaining the regular functions and activities of cells.
Viruses can trigger apoptosis of infected cells via a range of mechanisms including:
- Receptor binding
- Activation of protein kinase R (PKR)
- Interaction with p53
- Expression of viral proteins coupled to MHC proteins on the surface of the infected cell, allowing recognition by cells of the immune system (such as natural killer and cytotoxic T cells) that then induce the infected cell to undergo apoptosis.[105]
Canine distemper virus (CDV) is known to cause apoptosis in central nervous system and lymphoid tissue of infected dogs in vivo and in vitro.[106] Apoptosis caused by CDV is typically induced via the extrinsic pathway, which activates caspases that disrupt cellular function and eventually leads to the cells death.[90] In normal cells, CDV activates caspase-8 first, which works as the initiator protein followed by the executioner protein caspase-3.[90] However, apoptosis induced by CDV in HeLa cells does not involve the initiator protein caspase-8. HeLa cell apoptosis caused by CDV follows a different mechanism than that in vero cell lines.[90] This change in the caspase cascade suggests CDV induces apoptosis via the intrinsic pathway, excluding the need for the initiator caspase-8. The executioner protein is instead activated by the internal stimuli caused by viral infection not a caspase cascade.[90]
The Oropouche virus (OROV) is found in the family Bunyaviridae. The study of apoptosis brought on by Bunyaviridae was initiated in 1996, when it was observed that apoptosis was induced by the La Crosse virus into the kidney cells of baby hamsters and into the brains of baby mice.[107]
OROV is a disease that is transmitted between humans by the biting midge (Culicoides paraensis).[108] It is referred to as a zoonotic arbovirus and causes febrile illness, characterized by the onset of a sudden fever known as Oropouche fever.[109]
The Oropouche virus also causes disruption in cultured cells – cells that are cultivated in distinct and specific conditions. An example of this can be seen in HeLa cells, whereby the cells begin to degenerate shortly after they are infected.[107]
With the use of gel electrophoresis, it can be observed that OROV causes DNA fragmentation in HeLa cells. It can be interpreted by counting, measuring, and analyzing the cells of the Sub/G1 cell population.[107] When HeLA cells are infected with OROV, the cytochrome C is released from the membrane of the mitochondria, into the cytosol of the cells. This type of interaction shows that apoptosis is activated via an intrinsic pathway.[104]
In order for apoptosis to occur within OROV, viral uncoating, viral internalization, along with the replication of cells is necessary. Apoptosis in some viruses is activated by extracellular stimuli. However, studies have demonstrated that the OROV infection causes apoptosis to be activated through intracellular stimuli and involves the mitochondria.[107]
Many viruses encode proteins that can inhibit apoptosis.[110] Several viruses encode viral homologs of Bcl-2. These homologs can inhibit proapoptotic proteins such as BAX and BAK, which are essential for the activation of apoptosis. Examples of viral Bcl-2 proteins include the Epstein-Barr virus BHRF1 protein and the adenovirus E1B 19K protein.[111] Some viruses express caspase inhibitors that inhibit caspase activity and an example is the CrmA protein of cowpox viruses. Whilst a number of viruses can block the effects of TNF and Fas. For example, the M-T2 protein of myxoma viruses can bind TNF preventing it from binding the TNF receptor and inducing a response.[112] Furthermore, many viruses express p53 inhibitors that can bind p53 and inhibit its transcriptional transactivation activity. As a consequence, p53 cannot induce apoptosis, since it cannot induce the expression of proapoptotic proteins. The adenovirus E1B-55K protein and the hepatitis B virus HBx protein are examples of viral proteins that can perform such a function.[113]
Viruses can remain intact from apoptosis in particular in the latter stages of infection. They can be exported in the apoptotic bodies that pinch off from the surface of the dying cell, and the fact that they are engulfed by phagocytes prevents the initiation of a host response. This favours the spread of the virus.[112] Prions can cause apoptosis in neurons.
Plants
Programmed cell death in plants has a number of molecular similarities to that of animal apoptosis, but it also has differences, notable ones being the presence of a cell wall and the lack of an immune system that removes the pieces of the dead cell. Instead of an immune response, the dying cell synthesizes substances to break itself down and places them in a vacuole that ruptures as the cell dies. Additionally, plants do not contain phagocytic cells, which are essential in the process of breaking down and removing apoptotic bodies.[114] Whether this whole process resembles animal apoptosis closely enough to warrant using the name apoptosis (as opposed to the more general programmed cell death) is unclear.[115][116]
Caspase-independent apoptosis
The characterization of the caspases allowed the development of caspase inhibitors, which can be used to determine whether a cellular process involves active caspases. Using these inhibitors it was discovered that cells can die while displaying a morphology similar to apoptosis without caspase activation.[117] Later studies linked this phenomenon to the release of AIF (apoptosis-inducing factor) from the mitochondria and its translocation into the nucleus mediated by its NLS (nuclear localization signal). Inside the mitochondria, AIF is anchored to the inner membrane. In order to be released, the protein is cleaved by a calcium-dependent calpain protease.
See also
- Anoikis
- Apaf-1
- Apo2.7
- Apoptotic DNA fragmentation
- Atromentin induces apoptosis in human leukemia U937 cells.[118]
- Autolysis
- Autophagy
- Cisplatin
- Cytotoxicity
- Entosis
- Ferroptosis
- Homeostasis
- Immunology
- Necrobiosis
- Necrosis
- Necrotaxis
- Nemosis
- Mitotic catastrophe
- p53
- Paraptosis
- Pseudoapoptosis
- PI3K/AKT/mTOR pathway
Explanatory footnotes
- ↑ Note that the average human adult has more than 13 trillion cells (1.3×1013),[3] of which at most only 70 billion (7.0×1010) die per day. That is, about 5 out of every 1,000 cells (0.5%) die each day due to apoptosis.
- ↑ HeLa cells are an immortalized cancer cell line used frequently in research. The cell line was established by removing cells directly from Henrietta Lacks, a cancer patient.
Citations
- ↑ Means to an End: Apoptosis and other Cell Death Mechanisms. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. 2011. ISBN 978-0-87969-888-1. https://books.google.com/books?id=s8jBcQAACAAJ. Retrieved 2020-05-25.
- ↑ 2.0 2.1 "Apoptosis: the complex scenario for a silent cell death". Mol Imaging Biol 5 (1): 2–14. 2003. doi:10.1016/S1536-1632(03)00024-6. PMID 14499155.
- ↑ Alberts, p. 2.
- ↑ Apoptosis in Carcinogenesis and Chemotherapy. Netherlands: Springer. 2009. ISBN 978-1-4020-9597-9.
- ↑ "Chapter 18 Apoptosis: Programmed Cell Death Eliminates Unwanted Cells". Molecular Biology of the Cell (textbook) (5th ed.). Garland Science. 2008. p. 1115. ISBN 978-0-8153-4105-5.
- ↑ "A minimal model of signaling network elucidates cell-to-cell stochastic variability in apoptosis". PLOS ONE 5 (8): e11930. August 2010. doi:10.1371/journal.pone.0011930. PMID 20711445. Bibcode: 2010PLoSO...511930R.
- ↑ "Apoptosis: A Review of Programmed Cell Death". Toxicologic Pathology 35 (4): 495–516. June 2007. doi:10.1080/01926230701320337. PMID 17562483.
- ↑ "A histochemical study of hypertrophy and ischaemic injury of rat liver with special reference to changes in lysosomes". The Journal of Pathology and Bacteriology 90 (2): 419–435. October 1965. doi:10.1002/path.1700900210. PMID 5849603.
- ↑ Agency for Science, Technology and Research. "Prof Andrew H. Wyllie – Lecture Abstract". http://www.a-star.edu.sg/astar/biomed/action/biomed_dvp_abstract.do?id=2901ddeb02dH.
- ↑ 10.0 10.1 "Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics". British Journal of Cancer 26 (4): 239–257. August 1972. doi:10.1038/bjc.1972.33. PMID 4561027.
- ↑ "John Kerr and apoptosis". The Medical Journal of Australia 173 (11–12): 616–617. 2000. doi:10.5694/j.1326-5377.2000.tb139362.x. PMID 11379508.
- ↑ "Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells". Nature 335 (6189): 440–2. September 1988. doi:10.1038/335440a0. PMID 3262202. Bibcode: 1988Natur.335..440V.
- ↑ Alberts, p. 1021.
- ↑ 14.0 14.1 "The American Heritage Dictionary entry: apoptosis". Houghton Mifflin Harcourt Publishing Company. 2020. https://ahdictionary.com/word/search.html?q=apoptosis.
- ↑ Apoptosis Interest Group (1999). "About apoptosis". http://www.nih.gov/sigs/aig/Aboutapo.html.
- ↑ "Definition of apoptosis". http://www.webster.com/dictionary/apoptosis.
- ↑ Alberts, p. 1029.
- ↑ Alberts, p. 1023.
- ↑ Alberts, p. 1032.
- ↑ Alberts, p. 1024.
- ↑ Nirmala GJ and Lopus M (2020) Cell death mechanisms in eukaryotes. Cell Biol Toxicol, 36, 145–164. doi: /10.1007/s10565-019-09496-2. PMID 31820165
- ↑ 22.0 22.1 22.2 22.3 22.4 22.5 22.6 Robbins Pathologic Basis of Disease. Philadelphia: W.B Saunders Company. 1998. ISBN 978-0-7216-7335-6.
- ↑ "Saturated fatty acid-induced apoptosis in MDA-MB-231 breast cancer cells. A role for cardiolipin". The Journal of Biological Chemistry 278 (34): 31861–31870. August 2003. doi:10.1074/jbc.m300190200. PMID 12805375.
- ↑ "Calcium orchestrates apoptosis". Nature Cell Biology 5 (12): 1041–1043. December 2003. doi:10.1038/ncb1203-1041. PMID 14647298. https://zenodo.org/record/1233353. Retrieved 2018-05-18.
- ↑ "Selenium modulates oxidative stress-induced cell apoptosis in human myeloid HL-60 cells through regulation of calcium release and caspase-3 and -9 activities". The Journal of Membrane Biology 232 (1–3): 15–23. December 2009. doi:10.1007/s00232-009-9212-2. PMID 19898892.
- ↑ "Cell biology. PARP-1--a perpetrator of apoptotic cell death?". Science 297 (5579): 200–201. July 2002. doi:10.1126/science.1074592. PMID 12114611.
- ↑ "The coordinate release of cytochrome c during apoptosis is rapid, complete and kinetically invariant". Nature Cell Biology 2 (3): 156–162. March 2000. doi:10.1038/35004029. PMID 10707086.
- ↑ "Real-Time dynamics of Ca2+, caspase-3/7, and morphological changes in retinal ganglion cell apoptosis under elevated pressure". PLOS ONE 5 (10): e13437. October 2010. doi:10.1371/journal.pone.0013437. PMID 20976135. Bibcode: 2010PLoSO...513437L.
- ↑ "Role of Calcium Signals on Hydrogen Peroxide-Induced Apoptosis in Human Myeloid HL-60 Cells". International Journal of Biomedical Science 5 (3): 246–256. September 2009. doi:10.59566/IJBS.2009.5246. PMID 23675144.
- ↑ "Oxidative Stress-Induced Caspases are Regulated in Human Myeloid HL-60 Cells by Calcium Signal". Current Signal Transduction Therapy 5 (2): 181–186. 2010. doi:10.2174/157436210791112172.
- ↑ "Nitric oxide: NO apoptosis or turning it ON?". Cell Death and Differentiation 10 (8): 864–869. August 2003. doi:10.1038/sj.cdd.4401261. PMID 12867993.
- ↑ "Nitric oxide (NO): an effector of apoptosis". Cell Death and Differentiation 6 (10): 969–975. October 1999. doi:10.1038/sj.cdd.4400582. PMID 10556974.
- ↑ "Pore formation by dimeric Bak and Bax: an unusual pore?". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 372 (1726): 20160218. August 2017. doi:10.1098/rstb.2016.0218. PMID 28630157.
- ↑ "Structural biology. Controlling the caspases". Science 294 (5546): 1477–1478. November 2001. doi:10.1126/science.1062236. PMID 11711663.
- ↑ 35.0 35.1 "The Fas signaling pathway: more than a paradigm". Science 296 (5573): 1635–1636. May 2002. doi:10.1126/science.1071553. PMID 12040174. Bibcode: 2002Sci...296.1635W.
- ↑ "TNF-R1 signaling: a beautiful pathway". Science 296 (5573): 1634–1635. May 2002. doi:10.1126/science.1071924. PMID 12040173. Bibcode: 2002Sci...296.1634C.
- ↑ "Connection Map for Tumor Necrosis Factor Pathway". Science's STKE 2007 (382): tw132. 2007. doi:10.1126/stke.3822007tw132. http://stke.sciencemag.org/cgi/cm/CMP_7107. Retrieved 2004-01-01.
- ↑ "The lysosome-associated apoptosis-inducing protein containing the pleckstrin homology (PH) and FYVE domains (LAPF), representative of a novel family of PH and FYVE domain-containing proteins, induces caspase-independent apoptosis via the lysosomal-mitochondrial pathway". The Journal of Biological Chemistry 280 (49): 40985–40995. December 2005. doi:10.1074/jbc.M502190200. PMID 16188880.
- ↑ "Apoptosis in the development and treatment of cancer". Carcinogenesis 26 (2): 263–270. February 2005. doi:10.1093/carcin/bgh283. PMID 15375012.
- ↑ "Design and synthesis of a luminescent iridium complex-peptide hybrid (IPH) that detects cancer cells and induces their apoptosis". Bioorganic & Medicinal Chemistry 26 (17): 4804–4816. September 2018. doi:10.1016/j.bmc.2018.08.016. PMID 30177492.
- ↑ "Connection Map for Fas Signaling Pathway". Science's STKE 2007 (380): tr1. 2007. doi:10.1126/stke.3802007tr1. http://stke.sciencemag.org/cgi/cm/CMP_7966. Retrieved 2004-01-01.
- ↑ "Bcl-2 inhibits Bax translocation from cytosol to mitochondria during drug-induced apoptosis of human tumor cells". Cell Death and Differentiation 7 (1): 102–111. January 2000. doi:10.1038/sj.cdd.4400597. PMID 10713725.
- ↑ "Building blocks of the apoptotic pore: how Bax and Bak are activated and oligomerize during apoptosis". Cell Death and Differentiation 21 (2): 196–205. February 2014. doi:10.1038/cdd.2013.139. PMID 24162660.
- ↑ "Molecular characterization of mitochondrial apoptosis-inducing factor". Nature 397 (6718): 441–446. February 1999. doi:10.1038/17135. PMID 9989411. Bibcode: 1999Natur.397..441S.
- ↑ "Optogenetic Control of Apoptosis in Targeted Tissues of Xenopus laevis Embryos". Journal of Cell Death 7: 25–31. 2014. doi:10.4137/JCD.S18368. PMID 25374461.
- ↑ "Evolutionary Significance of Iodine". Current Chemical Biology 5 (3): 155–62. 2011. doi:10.2174/187231311796765012.
- ↑ Venturi, Sebastiano (2014). "Iodine, PUFAs and Iodolipids in Health and Disease: An Evolutionary Perspective". Human Evolution 29 (1–3): 185–205. ISSN 0393-9375.
- ↑ "Apoptosis and differentiation of Xenopus tail-derived myoblasts by thyroid hormone". Journal of Molecular Endocrinology 54 (3): 185–192. June 2015. doi:10.1530/JME-14-0327. PMID 25791374.
- ↑ 49.0 49.1 "Understanding Apoptosis and Apoptotic Pathways Targeted Cancer Therapeutics". Advanced Pharmaceutical Bulletin 9 (2): 205–218. June 2019. doi:10.15171/apb.2019.024. PMID 31380246.
- ↑ "BCL-2 family proteins: changing partners in the dance towards death". Cell Death and Differentiation 25 (1): 65–80. January 2018. doi:10.1038/cdd.2017.186. PMID 29149100.
- ↑ "Negative regulators of cell death pathways in cancer: perspective on biomarkers and targeted therapies". Apoptosis 23 (2): 93–112. February 2018. doi:10.1007/s10495-018-1440-4. PMID 29322476.
- ↑ "Apoptosis Triggers Specific, Rapid, and Global mRNA Decay with 3' Uridylated Intermediates Degraded by DIS3L2". Cell Reports 11 (7): 1079–1089. May 2015. doi:10.1016/j.celrep.2015.04.026. PMID 25959823.
- ↑ "Disruption of the cytoskeleton after apoptosis induction with autoantibodies". Autoimmunity 36 (3): 183–189. May 2003. doi:10.1080/0891693031000105617. PMID 12911286.
- ↑ "Two distinct pathways leading to nuclear apoptosis". The Journal of Experimental Medicine 192 (4): 571–580. August 2000. doi:10.1084/jem.192.4.571. PMID 10952727.
- ↑ "Sequential degradation of proteins from the nuclear envelope during apoptosis". Journal of Cell Science 114 (Pt 20): 3643–3653. October 2001. doi:10.1242/jcs.114.20.3643. PMID 11707516.
- ↑ "Apoptotic DNA fragmentation". Experimental Cell Research 256 (1): 12–18. April 2000. doi:10.1006/excr.2000.4834. PMID 10739646.
- ↑ "A selective procedure for DNA extraction from apoptotic cells applicable for gel electrophoresis and flow cytometry". Analytical Biochemistry 218 (2): 314–319. May 1994. doi:10.1006/abio.1994.1184. PMID 8074286.
- ↑ "An evaluation of renal tubular DNA laddering in response to oxygen deprivation and oxidant injury". Journal of the American Society of Nephrology 5 (6): 1307–1313. December 1994. doi:10.1681/ASN.V561307. PMID 7893995.
- ↑ "Cell disassembly during apoptosis" (in en). WikiJournal of Medicine 4 (1). 2017. doi:10.15347/wjm/2017.008.
- ↑ 60.0 60.1 "Defining the morphologic features and products of cell disassembly during apoptosis". Apoptosis 22 (3): 475–477. March 2017. doi:10.1007/s10495-017-1345-7. PMID 28102458.
- ↑ "Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I". Nature Cell Biology 3 (4): 339–345. April 2001. doi:10.1038/35070009. PMID 11283606.
- ↑ "Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing". Nature Cell Biology 3 (4): 346–352. April 2001. doi:10.1038/35070019. PMID 11283607.
- ↑ "A novel role for microtubules in apoptotic chromatin dynamics and cellular fragmentation". Journal of Cell Science 119 (Pt 11): 2362–2374. June 2006. doi:10.1242/jcs.02959. PMID 16723742.
- ↑ 64.0 64.1 "Unexpected link between an antibiotic, pannexin channels and apoptosis". Nature 507 (7492): 329–334. March 2014. doi:10.1038/nature13147. PMID 24646995. Bibcode: 2014Natur.507..329P.
- ↑ "A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure". Nature Communications 6: 7439. June 2015. doi:10.1038/ncomms8439. PMID 26074490. Bibcode: 2015NatCo...6.7439A.
- ↑ "Burying the dead: the impact of failed apoptotic cell removal (efferocytosis) on chronic inflammatory lung disease". Chest 129 (6): 1673–1682. June 2006. doi:10.1378/chest.129.6.1673. PMID 16778289.
- ↑ "Phosphatidylserine receptor is required for clearance of apoptotic cells". Science 302 (5650): 1560–1563. November 2003. doi:10.1126/science.1087621. PMID 14645847. Bibcode: 2003Sci...302.1560O.
- ↑ "Cell corpse engulfment mediated by C. elegans phosphatidylserine receptor through CED-5 and CED-12". Science 302 (5650): 1563–1566. November 2003. doi:10.1126/science.1087641. PMID 14645848. Bibcode: 2003Sci...302.1563W. http://ntur.lib.ntu.edu.tw/handle/246246/161415. Retrieved 2017-02-26.
- ↑ "Cell biology. Eat me or die". Science 302 (5650): 1516–1517. November 2003. doi:10.1126/science.1092533. PMID 14645835.
- ↑ Phagocytosis of dying cells: from molecular mechanisms to human diseases. Springer. 2009-01-14. doi:10.1007/978-1-4020-9293-0. ISBN 978-1-4020-9292-3. https://www.springer.com/biomed/cancer/book/978-1-4020-9292-3. Retrieved 2017-08-28.
- ↑ "Segregation of RNA and separate packaging of DNA and RNA in apoptotic bodies during apoptosis". Experimental Cell Research 260 (2): 248–256. November 2000. doi:10.1006/excr.2000.5027. PMID 11035919.
- ↑ "The role of mitochondria in apoptosis". Annual Review of Genetics 43 (1): 95–118. 2009-12-01. doi:10.1146/annurev-genet-102108-134850. PMID 19659442.
- ↑ "Characterization of tumor necrosis factor-deficient mice". Proceedings of the National Academy of Sciences of the United States of America 94 (15): 8093–8098. July 1997. doi:10.1073/pnas.94.15.8093. PMID 9223320. Bibcode: 1997PNAS...94.8093M.
- ↑ "Regulation of the Apaf-1-caspase-9 apoptosome". Journal of Cell Science 123 (Pt 19): 3209–3214. October 2010. doi:10.1242/jcs.073643. PMID 20844150.
- ↑ "A common pathway mediates retinoic acid and PMA-dependent programmed cell death (apoptosis) of neuronal cells". Brain Research 644 (1): 7–12. April 1994. doi:10.1016/0006-8993(94)90339-5. PMID 8032951.
- ↑ "A common pathway mediates retinoic acid and PMA-dependent programmed cell death (apoptosis) of neuronal cells". Brain Research 644 (1): 7–12. April 1994. doi:10.1016/0006-8993(94)90339-5. PMID 8032951.
- ↑ "Density gradient capacitation is the most suitable method to improve fertilization and to reduce DNA fragmentation positive spermatozoa of infertile men". Anatolian Journal of Obstetrics & Gynecology 3 (1): 1–7. 2009. https://www.researchgate.net/publication/259983025. Retrieved 2016-03-08.
- ↑ "Cytometry in cell necrobiology: analysis of apoptosis and accidental cell death (necrosis)". Cytometry 27 (1): 1–20. January 1997. doi:10.1002/(sici)1097-0320(19970101)27:1<1::aid-cyto2>3.0.co;2-l. PMID 9000580.
- ↑ "Chapter 16 Methods for Distinguishing Apoptotic from Necrotic Cells and Measuring Their Clearance". Programmed Cell Death, General Principles for Studying Cell Death, Part A. Methods in Enzymology. 442. 2008. pp. 307–41. doi:10.1016/S0076-6879(08)01416-X. ISBN 9780123743121.
- ↑ "Apoptosis and necrosis: detection, discrimination and phagocytosis". Methods 44 (3): 205–221. March 2008. doi:10.1016/j.ymeth.2007.12.001. PMID 18314051.
- ↑ "Determination of apoptotic and necrotic cell death in vitro and in vivo". Methods 61 (2): 117–129. June 2013. doi:10.1016/j.ymeth.2013.02.011. PMID 23473780. https://zenodo.org/record/3423520. Retrieved 2019-11-05.
- ↑ "Apoptosis and beyond: cytometry in studies of programmed cell death". Recent Advances in Cytometry, Part B - Advances in Applications. Methods in Cell Biology. 103. 2011. pp. 55–98. doi:10.1016/B978-0-12-385493-3.00004-8. ISBN 9780123854933.
- ↑ "Apoptosis in the pathogenesis and treatment of disease". Science 267 (5203): 1456–1462. March 1995. doi:10.1126/science.7878464. PMID 7878464. Bibcode: 1995Sci...267.1456T.
- ↑ "Predominant suppression of apoptosome by inhibitor of apoptosis protein in non-small cell lung cancer H460 cells: therapeutic effect of a novel polyarginine-conjugated Smac peptide". Cancer Research 63 (4): 831–837. February 2003. PMID 12591734. http://cancerres.aacrjournals.org/cgi/pmidlookup?view=long&pmid=12591734. Retrieved 2008-09-04.
- ↑ "Aberrant control of NF-κB in cancer permits transcriptional and phenotypic plasticity, to curtail dependence on host tissue: molecular mode". Cancer Biology & Medicine 14 (3): 254–270. August 2017. doi:10.20892/j.issn.2095-3941.2017.0029. PMID 28884042.
- ↑ "Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence". Nature 424 (6948): 516–523. July 2003. doi:10.1038/nature01850. PMID 12872134. Bibcode: 2003Natur.424..516T.
- ↑ "DNA repair/pro-apoptotic dual-role proteins in five major DNA repair pathways: fail-safe protection against carcinogenesis". Mutation Research 511 (2): 145–178. June 2002. doi:10.1016/S1383-5742(02)00009-1. PMID 12052432.
- ↑ 88.0 88.1 "Apoptosis: its origin, history, maintenance and the medical implications for cancer and aging". Physical Biology 13 (3): 031001. May 2016. doi:10.1088/1478-3975/13/3/031001. PMID 27172135. Bibcode: 2016PhBio..13c1001K. http://eprints.ibb.waw.pl/1615/1/Apoptosis2016.pdf. Retrieved 2019-12-26.
- ↑ "On the origin of cancer cells". Science 123 (3191): 309–314. February 1956. doi:10.1126/science.123.3191.309. PMID 13298683. Bibcode: 1956Sci...123..309W.
- ↑ 90.0 90.1 90.2 90.3 90.4 90.5 "Canine distemper virus induces apoptosis in cervical tumor derived cell lines". Virology Journal 8 (1): 334. June 2011. doi:10.1186/1743-422X-8-334. PMID 21718481.
- ↑ "Inhibition of apoptosis in human laryngeal cancer cells by E6 and E7 oncoproteins of human papillomavirus 16". Journal of Cellular Biochemistry 103 (4): 1125–1143. March 2008. doi:10.1002/jcb.21490. PMID 17668439.
- ↑ 92.0 92.1 "Inhibition of HPV 16 E6 oncogene expression by RNA interference in vitro and in vivo". International Journal of Gynecological Cancer 16 (2): 743–751. 2006. doi:10.1111/j.1525-1438.2006.00384.x. PMID 16681755.
- ↑ "Human papillomavirus type 16 E6 and HPV-16 E6/E7 sensitize human keratinocytes to apoptosis induced by chemotherapeutic agents: roles of p53 and caspase activation". Journal of Cellular Biochemistry 78 (2): 334–349. May 2000. doi:10.1002/(sici)1097-4644(20000801)78:2<334::aid-jcb15>3.3.co;2-6. PMID 10842327.
- ↑ "Apoptosis in physiological and pathological skin: implications for therapy". Current Molecular Medicine 6 (4): 375–394. June 2006. doi:10.2174/156652406777435390. PMID 16900661.
- ↑ "The Alzheimer's A beta peptide induces neurodegeneration and apoptotic cell death in transgenic mice". Nature Genetics 9 (1): 21–30. January 1995. doi:10.1038/ng0195-21. PMID 7704018.
- ↑ "Histochemical detection of apoptosis in Parkinson's disease". Journal of the Neurological Sciences 137 (2): 120–123. May 1996. doi:10.1016/0022-510X(95)00336-Z. PMID 8782165.
- ↑ "Alzheimer's disease: the amyloid hypothesis and the Inverse Warburg effect". Frontiers in Physiology 5: 522. 2014. doi:10.3389/fphys.2014.00522. PMID 25642192.
- ↑ "Inverse occurrence of cancer and Alzheimer disease: a population-based incidence study". Neurology 81 (4): 322–328. July 2013. doi:10.1212/WNL.0b013e31829c5ec1. PMID 23843468.
- ↑ "Apoptosis induction by a novel retinoid-related molecule requires nuclear factor-kappaB activation". Cancer Research 65 (11): 4909–4917. June 2005. doi:10.1158/0008-5472.CAN-04-4124. PMID 15930313.
- ↑ "Mechanisms of CD4+ T lymphocyte cell death in human immunodeficiency virus infection and AIDS". The Journal of General Virology 84 (Pt 7): 1649–1661. July 2003. doi:10.1099/vir.0.19110-0. PMID 12810858.
- ↑ "HIV-1 expression induces tubular cell G2/M arrest and apoptosis". Renal Failure 30 (6): 655–664. 2008. doi:10.1080/08860220802134672. PMID 18661417.
- ↑ Indiana University Health. "AIDS Defining Criteria | Riley". IU Health. http://iuhealth.org/riley/infectious-diseases/hiv/aids-defining-criteria/.
- ↑ "A clue to unprecedented strategy to HIV eradication: "Lock-in and apoptosis"". Scientific Reports 7 (1): 8957. August 2017. doi:10.1038/s41598-017-09129-w. PMID 28827668. Bibcode: 2017NatSR...7.8957T.
- ↑ 104.0 104.1 "Recent advances in apoptosis, mitochondria and drug resistance in cancer cells". Biochimica et Biophysica Acta (BBA) - Bioenergetics 1807 (6): 735–745. June 2011. doi:10.1016/j.bbabio.2011.03.010. PMID 21453675.
- ↑ "Apoptosis: an innate immune response to virus infection". Trends in Microbiology 7 (4): 160–165. April 1999. doi:10.1016/S0966-842X(99)01487-0. PMID 10217831.
- ↑ "Involvement of apoptosis in syncytial cell death induced by canine distemper virus". Comparative Immunology, Microbiology and Infectious Diseases 27 (6): 445–455. November 2004. doi:10.1016/j.cimid.2004.01.007. PMID 15325517.
- ↑ 107.0 107.1 107.2 107.3 "Apoptosis induced by Oropouche virus infection in HeLa cells is dependent on virus protein expression". Virus Research 149 (1): 56–63. April 2010. doi:10.1016/j.virusres.2009.12.013. PMID 20080135.
- ↑ "Reemergence of Oropouche fever, northern Brazil". Emerging Infectious Diseases 13 (6): 912–915. June 2007. doi:10.3201/eid1306.061114. PMID 17553235.
- ↑ "Oropouche virus entry into HeLa cells involves clathrin and requires endosomal acidification". Virus Research 138 (1–2): 139–143. December 2008. doi:10.1016/j.virusres.2008.08.016. PMID 18840482.
- ↑ "Regulation of apoptosis by viral gene products". Journal of Virology 71 (3): 1739–1746. March 1997. doi:10.1128/jvi.71.3.1739-1746.1997. PMID 9032302.
- ↑ "Viral Bcl-2 homologs and their role in virus replication and associated diseases". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1644 (2–3): 211–227. March 2004. doi:10.1016/j.bbamcr.2003.11.001. PMID 14996505.
- ↑ 112.0 112.1 "A time to kill: viral manipulation of the cell death program". The Journal of General Virology 83 (Pt 7): 1547–1564. July 2002. doi:10.1099/0022-1317-83-7-1547. PMID 12075073.
- ↑ "Abrogation of p53-induced apoptosis by the hepatitis B virus X gene". Cancer Research 55 (24): 6012–6016. December 1995. PMID 8521383.
- ↑ "Morphological classification of plant cell deaths". Cell Death and Differentiation 18 (8): 1241–1246. August 2011. doi:10.1038/cdd.2011.36. PMID 21494263.
- ↑ "Programmed cell death in plants resembles apoptosis of animals". Biotecnología Aplicada 23: 1–10. 2006. http://elfosscientiae.cigb.edu.cu/PDFs/BA/2006/23/1/BA002301RV001-010.pdf.
- ↑ "Reassessing apoptosis in plants". Nature Plants 3 (10): 773–779. October 2017. doi:10.1038/s41477-017-0020-x. PMID 28947814.
- ↑ "BAX-induced cell death may not require interleukin 1 beta-converting enzyme-like proteases". Proceedings of the National Academy of Sciences of the United States of America 93 (25): 14559–14563. December 1996. doi:10.1073/pnas.93.25.14559. PMID 8962091. Bibcode: 1996PNAS...9314559X.
- ↑ "Atromentin-induced apoptosis in human leukemia U937 cells". Journal of Microbiology and Biotechnology 19 (9): 946–950. September 2009. doi:10.4014/jmb.0811.617. PMID 19809251.
General bibliography
- Molecular Biology of the Cell (6th ed.). Garland Science. 2015. p. 2. ISBN 978-0815344322.
External links
- Apoptosis & cell surface[yes|permanent dead link|dead link}}]
- Apoptosis & Caspase 3, The Proteolysis Map – animation
- Apoptosis & Caspase 8, The Proteolysis Map – animation
- Apoptosis & Caspase 7, The Proteolysis Map – animation
- Apoptosis MiniCOPE Dictionary – list of apoptosis terms and acronyms
- Apoptosis (Programmed Cell Death) – The Virtual Library of Biochemistry, Molecular Biology and Cell Biology
- Apoptosis Research Portal
- Apoptosis Info Apoptosis protocols, articles, news, and recent publications.
- Database of proteins involved in apoptosis
- Apoptosis Video
- Apoptosis Video (WEHI on YouTube )
- The Mechanisms of Apoptosis Kimball's Biology Pages. Simple explanation of the mechanisms of apoptosis triggered by internal signals (bcl-2), along the caspase-9, caspase-3 and caspase-7 pathway; and by external signals (FAS and TNF), along the caspase 8 pathway. Accessed 25 March 2007.
- WikiPathways – Apoptosis pathway
- "Finding Cancer's Self-Destruct Button". CR magazine (Spring 2007). Article on apoptosis and cancer.
- Xiaodong Wang's lecture: Introduction to Apoptosis
- Robert Horvitz's Short Clip: Discovering Programmed Cell Death
- The Bcl-2 Database
- DeathBase: a database of proteins involved in cell death, curated by experts
- European Cell Death Organization
- Apoptosis signaling pathway created by Cusabio
 |