Biology:Gap junction
| Gap junction | |
|---|---|
 Vertebrate gap junction | |
| Anatomical terminology |
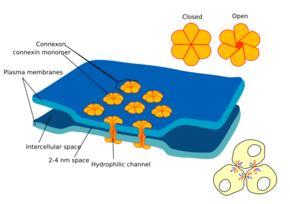
Gap junctions are membrane channels between adjacent cells that allow the direct exchange of cytoplasmic substances, such as small molecules, substrates, and metabolites.[1]
Gap junctions were first described as close appositions alongside tight junctions, however, electron microscopy studies in 1967 led to gap junctions being named as such to be distinguished from tight junctions.[2] They bridge a 2-4 nm gap between cell membranes.[3]
Gap junctions use protein complexes known as connexons, composed of connexin proteins to connect one cell to another. Gap junction proteins include the more than 26 types of connexin, as well as at least 12 non-connexin components that make up the gap junction complex or nexus,[4] including the tight junction protein ZO-1—a protein that holds membrane content together and adds structural clarity to a cell,[5] sodium channels,[6] and aquaporin.[7][8]
More gap junction proteins have become known due to the development of next-generation sequencing. Connexins were found to be structurally homologous between vertebrates and invertebrates but different in sequence.[9] As a result, the term innexin is used to differentiate invertebrate connexins.[10] There are more than 20 known innexins,[11] along with unnexins in parasites and vinnexins in viruses.
An electrical synapse is a gap junction that can transmit action potentials between neurons. Such synapses create bidirectional continuous-time electrical coupling[12][13] between neurons. Connexon pairs act as generalized regulated gates for ions and smaller molecules between cells. Hemichannel connexons form channels to the extracellular environment.[14][15][16][17]
A gap junction or macula communicans is different from an ephaptic coupling that involves electrical signals external to the cells.[18][19]
Structure

In vertebrates, gap junction hemichannels are primarily homo- or hetero-hexamers of connexin proteins. Hetero-hexamers at gap junction plaques, help form a uniform intercellular space of 2-4 nm.[21] In this way hemichannels in the membrane of each cell are aligned with one another forming an intercellular communication path.[22]
Invertebrate gap junctions comprise proteins from the innexin family. Innexins have no significant sequence homology with connexins.[23] Though differing in sequence to connexins, innexins are similar enough to connexins to form gap junctions in vivo in the same way connexins do.[24][25][26]
The more recently characterized pannexin family,[27] which was originally thought to form intercellular channels (with an amino acid sequence similar to innexins[28]) in fact functions as a single-membrane channel that communicates with the extracellular environment and has been shown to pass calcium and ATP.[29] This has led to the idea that pannexins may not form intercellular junctions in the same way connexins and innexins do and therefore should not use the same hemi-channel/channel naming.[30] Others have presented evidence based on genetic sequencing and overall functioning in tissues, that pannexins should still be considered part of the gap junction family of proteins despite structural differences. These researchers also note that there are still more groups of connexin orthologs to be discovered.[31]
Gap junction channels formed from two identical hemichannels are called homotypic, while those with differing hemichannels are heterotypic. In turn, hemichannels of uniform protein composition are called homomeric, while those with differing proteins are heteromeric. Channel composition influences the function of gap junction channels, and different connexins will not necessarily form heterotypic with all others.[32]
Before innexins and connexins were well characterized, the genes coding for the connexin gap junction channels were classified in one of three groups (A, B and C; for example, GJA1, GJC1), based on gene mapping and sequence similarity.[33][34][35] However, connexin genes do not code directly for the expression of gap junction channels; genes can produce only the proteins that make up gap junction channels. An alternative naming system based on the protein's molecular weight is the most widely used (for example, connexin43=GJA1, connexin30.3=GJB4).
Levels of organization
In vertebrates, two pairs of six connexin proteins form a connexon. In invertebrates, six innexin proteins form an innexon. Otherwise, the structures are similar.
- The connexin genes (DNA) are transcribed to RNA, which is then translated to produce a connexin.
- One connexin protein has four transmembrane domains[21][36]
- Six connexin proteins create one connexon channel a hemichannel. When identical connexin proteins join to form one connexon, it is called a homomeric connexon. When different connexin proteins join to form one connexon, it is called a heteromeric connexon.
- Two connexons, joined across a cell membrane, comprise a gap junction channel.
When two identical connexons come together to form a gap junction channel, it is called a homotypic channel. When one homomeric connexon and one heteromeric connexon come together, it is called a heterotypic gap junction channel. When two heteromeric connexons join, it is also called a heterotypic gap junction channel. - Tens to thousands of gap junction channels cluster in areas to enable connexon pairs to form.[37] The macromolecular complex is called a gap junction plaque. Molecules other than connexins are involved in gap junction plaques including tight junction protein 1 and sodium channels.[5][6]
Properties of connexon pairs

A connexon or innexon channel pair:
- Allows for direct electrical communication between cells, although different hemichannel subunits can impart different single channel conductances, from about 30 pS to 500 pS.
- Allows for chemical communication between cells through the transmission of small second messengers, such as inositol triphosphate (IP3) and calcium (Ca2+),[39] although different hemichannel subunits can impart different selectivities for particular molecules.
- Generally allows transmembrane movement of molecules smaller than 485 daltons[40] (1,100 daltons through invertebrate gap junctions[41]), although different hemichannel subunits may impart different pore sizes and different charge selectivity. Large biomolecules, including nucleic acids and proteins, are precluded from cytoplasmic transfer between cells through gap junction hemichannel pairs.
- Ensures that molecules and current passing through the gap junction do not leak into the intercellular space.
Properties of connexons as hemichannels
Unpaired connexons or innexons can act as hemichannels in a single membrane, allowing the cell to exchange molecules directly with the exterior of the cell. It has been shown that connexons would be available to do this prior to being incorporated into the gap junction plaques.[37] Some of the properties of these unpaired connexons are listed below:
- Pore or transmembrane channel size is highly variable, in the range of approximately 8-20Å in diameter.[42]
- They connect the cytoplasm of the cell to the cell exterior and are thought to be in a closed state by default in order to prevent leakage from the cell.[43][44]
- Some connexons respond to external factors by opening up. Mechanical shear and various diseases can cause this to happen.[45]
Establishing further connexon properties different to those of connexon pairs, proves difficult due to separating their effects experimentally in organisms.[45]
Occurrence and distribution
Gap junctions have been observed in various animal organs and tissues where cells contact each other. From the 1950s to 1970s they were detected in:
- Human islet of Langerhans,[46] myometrium,[47] and eye lens[48]
- Rat pancreas, liver, adrenal cortex, epididymis, duodenum, muscle,[49] and seminiferous tubules[50]
- Rabbit cornea,[51] ovary,[52] and skin[53]
- Monkey retina[54]
- Chick embryos[55]
- Frog embryos[56]
- Fish blastoderm[57]
- Crayfish nerves[58]
- Lamprey and tunicate heart[59][60]
- Goldfish and hamster pressure-sensing acoustico-vestibular receptors[61]
- Daphnia hepatic caecum[62]
- Cephalopod digestive epithelium[63]
- Hydra muscle[64]
- Cockroach hemocyte capsules[65]
- Reaggregated cells[66][67]
Gap junctions have continued to be found in nearly all healthy animal cells that touch each other. Techniques such as confocal microscopy allow more rapid surveys of large areas of tissue. Tissues that were traditionally considered to have isolated cells such as in bone were shown to have cells that were still connected with gap junctions, however tenuously.[68] Exceptions to this are cells not normally in contact with neighboring cells, such as blood cells suspended in blood plasma. Adult skeletal muscle is a possible exception to the rule though their large size makes it difficult to be certain of this. An argument used against skeletal muscle gap junctions is that if they were present gap junctions may propagate contractions in an arbitrary way through cells making up the muscle. However, other muscle types do have gap junctions which do not cause arbitrary contractions.[69] Sometimes the number of gap junctions are reduced or absent in diseased tissues such as cancers[70][71][72] or the aging process.[73]
Since the discovery of innexins, pannexins and unnexins, gaps in our knowledge of intercellular communication are becoming more defined. Innexins look and behave similarly to connexins and can be seen to fill a similar role to connexins in invertebrates. Pannexins also look individually similar to connexins though they do not appear to easily form gap junctions. Of the over 20 metazoan groups connexins have been found only in vertebrata and tunicata. Innexins and pannexins are far more widespread including innexin homologues in vertebrates.[74][75] The unicellular Trypanosomatidae parasites presumably have unnexin genes to aid in their infection of animals including humans.[76] The even smaller adenovirus has its own vinnexin,[77] apparently derived from an innexin, to aid its transmission between the virus's insect hosts.
The term gap junction cannot be defined by a single protein or family of proteins with a specific function. For example, gap junction structures are found in sponges, despite the absence of pannexins. While we are still at the early stages of understanding the nervous system of a sponge[78] the gap junctions of sponges may as yet indicate intercellular communications pathways.[79][80]
Functions
At least five discrete functions have been ascribed to gap junction proteins:
- Electrical and metabolic coupling between cells
- Electrical and metabolic exchange through hemichannels
- Tumor suppressor genes (Cx43, Cx32 and Cx36)
- Adhesive function independent of conductive gap junction channel (neural migration in neocortex)
- Role of carboxyl-terminal in signaling cytoplasmic pathways (Cx43)
In a more general sense, gap junctions may be seen to function at the simplest level as a direct cell to cell pathway for electrical currents, small molecules and ions. The control of this communication allows complex downstream effects on multicellular organisms.
Embryonic, organ and tissue development
In the 1980s, more subtle roles of gap junctions in communication have been investigated. It was discovered that gap junction communication could be disrupted by adding anti-connexin antibodies into embryonic cells.[81][82] Embryos with areas of blocked gap junctions failed to develop normally. The mechanism by which antibodies blocked the gap junctions was unclear; systematic studies were undertaken to elucidate the mechanism.[83][84] Refinement of these studies suggested that gap junctions were key in the development of cell polarity[85] and the left-right symmetry in animals.[86][87] While signaling that determines the position of body organs appears to rely on gap junctions, so does the more fundamental differentiation of cells at later stages of embryonic development.[88][89][90][91][92]
Gap junctions were found to be responsible for the transmission of signals required for drugs to have an effect.[93] Conversely, some drugs were shown to block gap junction channels.[94]
The bystander effect and disease
Cell death
The bystander effect has its connotations of the innocent bystander being killed. When cells are dying or compromised due to disease or injury, messages are transmitted to neighboring cells by gap junctions. This can cause otherwise healthy bystander cells to also die.[95]
The bystander effect was later researched with regard to cells damaged by radiation or mechanical injury and in turn wound healing.[96][97][98][99][100] Disease seems to have an effect on the ability of gap junctions to fulfill their roles in wound healing.[101][102] The oral administration of gap junction blockers to reduce the symptoms of disease in remote parts of the body is slowly becoming a reality.[103]
Tissue restructuring
While there has been a tendency to focus on the bystander effect in disease due to the possibility of therapeutic avenues, there is evidence that there is a more central role in normal development of tissues. Death of some cells and their surrounding matrix may be required for a tissue to reach its final configuration; gap junctions appear essential to this process.[104][105] There are also more complex studies that try to combine our understanding of the simultaneous roles of gap junctions in both wound healing and tissue development.[106][107][108]
Disease
Mutations in connexins have been associated with many diseases in humans, including deafness,[109] heart atrial fibrillation (standstill) and cataracts. The study of these mutations has helped clarify some of the functions of connexins.[110][111]
Hemichannels are thought to play a general role in the progression and severity of many diseases; this is in part due to hemichannels being an open door to the outside of each cell.[45]
Areas of electrical coupling
Gap junctions electrically couple cells throughout the body of most animals. Electrical coupling can be relatively fast acting and can be used over short distances within an organism. Tissues in this section have well known functions observed to be coordinated by gap junctions, with intercellular signaling happening in time frames of microseconds or less.
Heart

Gap junctions are particularly important in cardiac muscle: the signal to contract is passed efficiently through gap junctions, allowing the heart muscle cells to contract in unison. The importance is emphasized by a secondary ephaptic pathway for the signal to contract also being associated with the gap junction plaques. This redundancy in signal transmission associated with gap junction plaques is the first to be described and involves sodium channels rather than connexins.[6][112]
Eye lens

Precise control of light refraction, structural dimensions and transparency are key aspects of the eye lens structure that allow focusing by the eye. Transparency is aided by the absence of nerves and blood vessels from the lens, so gap junctions are left with a larger loading of intercellular communication than in other tissues reflected in large numbers of gap junctions. The crystallinity of the lens also means the cells and gap junctions are well ordered for systematic mapping of where the gap junction plaques are. As no cells are lost from the lens interior during the life of the animal, a complete map of the gap junctions is possible.[113]
The associated figure shows how the size, shape, and frequency of gap junction plaques change with cell growth. With growth, fiber cells are progressively isolated from more direct metabolite exchange with the aqueous humor through the capsule and lens epithelium. The isolation correlates with the classical circular shape of larger plaques shown in the yellow zone being disrupted. Changing the fiber cells' morphology requires the movements of vesicles through the gap junction plaques at higher frequencies in this area.[114]
Neurons
A gap junction located between neurons is often referred to as an electrical synapse. The electrical synapse was discovered using electrical measurements before the gap junction structure was described. In mammals, electrical synapses are present throughout the central nervous system and have been studied specifically in the neocortex, hippocampus, vestibular nucleus, thalamic reticular nucleus, locus coeruleus, inferior olivary nucleus, mesencephalic nucleus of the trigeminal nerve, ventral tegmental area, olfactory bulb, retina and spinal cord of vertebrates.[115] In invertebrates, gap junctions are known to be expressed widely in the brain of the fruit fly, Drosophila.[116]
There has been some observation of coupling in the locus coeruleus between weak neurons and glial cells and in the cerebellum between Purkinje neurons and Bergmann glial cells. It appears that astrocytes are coupled by gap junctions, both to other astrocytes and to oligodendrocytes.[117] Moreover, mutations in the gap junction genes Cx43 and Cx56.6 cause white matter degeneration similar to that observed in Pelizaeus–Merzbacher disease and multiple sclerosis.
Connexin proteins expressed in neuronal gap junctions include mCX36, mCX57, and mCX45, with mRNAs for at least five other connexins (mCx26, mCx30.2, mCx32, mCx43, mCx47) detected but without immunocytochemical evidence for the corresponding protein within ultrastructurally defined gap junctions. Those mRNAs appear to be downregulated or destroyed by micro interfering RNAs (miRNAs) that are cell-type and cell-lineage specific.
Within the brain of the fruit fly Drosophila, gap junctions are known to be critical for a variety of functions.[118] [116] [119]
Astrocytes
An important feature of astrocytes is their high expression levels of the gap junction proteins connexin 30 (Cx30) and connexin 43 (Cx43). These proteins play crucial roles in regulating brain homeostasis through potassium buffering, intercellular communication, and nutrient transport.[120] Connexins typically form gap junction channels that allow direct intercellular communication between astrocytes. However, they can also form hemichannels that facilitate the exchange of ions and molecules with the extracellular space.
Studies have highlighted channel-independent functions of connexins, involving intracellular signaling, protein interactions, and cell adhesion.[121] Specifically, Cx30 has been shown to regulate the insertion of astroglial processes into synaptic clefts, which controls the efficacy of glutamate clearance. This, in turn, affects the synaptic strength and long-term plasticity of excitatory terminals, indicating a significant role in modulating synaptic transmission. Levels of Cx30 regulate synaptic glutamate concentration, hippocampal excitatory synaptic strength, plasticity, and memory. Astroglial networks have a physiologically optimized size to appropriately regulate neuronal functions.[122]
Cx30 is not limited to regulating excitatory synaptic transmission but also plays a crucial role in inhibitory synaptic regulation and broader neuronal network activities.[123] This highlights the importance of connexins in maintaining the intricate balance required for proper brain function.
Retina
Neurons within the retina show extensive coupling, both within populations of one cell type and between different cell types.[124]
Uterus
The uterine muscle (myometrium) remains in a quiescent relaxed state during pregnancy to maintain fetal development. Immediately preceding labor, the myometrium transforms into an activated contractile unit by increasing expression of connexin-43 (CX43, a.k.a. Gap Junction Alpha-1 protein, GJA1) facilitating gap junction (GJ) formation between individual myometrial cells. Importantly, the formation of GJs promotes communication between neighbouring myocytes, which facilitates the transfer of small molecules such as secondary messengers, metabolites, and small ions for electrical coupling. Consistent with all species, uterine myometrial contractions propagate from spontaneous action potentials as a result of sudden change in plasma membrane permeability. This leads to an increase of intracellular Ca²⁺ concentration, facilitating action potential propagation through electrically coupled cells.[125] It has more recently been discovered that uterine macrophages directly physically couples with uterine myocytes through CX43, transferring Ca²⁺, to promote uterine muscle contraction and excitation during labor onset.[126]
Hemichannel function
Hemichannels contribute to a cellular network of gap junctions and allow the release of adenosine triphosphate, glutamate, Nicotinamide adenine dinucleotide, and prostaglandin E2 from cells, which can all act as messengers to cells otherwise disconnected from such messaging.[127] In this sense, a gap junction plaque forms a one-to-one relationship with the neighboring cell, daisy chaining many cells together. Hemichannels form a one to many relationship with the surrounding tissue.
On a larger scale, the one-to-many communication of cells is typically carried out by the vascular and nervous systems. This makes detecting the contribution of hemichannels to extracellular communication more difficult in whole organisms. With the eye lens, the vascular and nervous systems are absent, making reliance on hemichannels greater and their detection easier. At the interface of the lens with the aqueous humor (where the lens exchanges metabolites), both gap junction plaques and more diffused connexon distribution can be seen in the accompanying micrographs.
Discovery
Form to function
Well before the demonstration of the gap in gap junctions, they were seen at the junction of neighboring nerve cells. The close proximity of the neighboring cell membranes at the gap junction led researchers to speculate that they had a role in intercellular communication, in particular the transmission of electrical signals.[58][128][129] Gap junctions were also found to be electrically rectifying in the early studies and referred to as an electrical synapse[130][131] but are now known to be bidirectional in general.[13][12] Later, it was found that chemicals could also be transported between cells through gap junctions.[132]
Implicit or explicit in most of the early studies is that the area of the gap junction was different in structure to the surrounding membranes in a way that made it look different. The gap junction had been shown to create a micro-environment between the two cells in the extracellular space or gap. This portion of extracellular space was somewhat isolated from the surrounding space and also bridged by what we now call connexon pairs, which form even more tightly sealed bridges that cross the gap junction gap between two cells. When viewed in the plane of the membrane by freeze-fracture techniques, higher-resolution distribution of connexons within the gap junction plaque is possible.[133]
Connexin free islands are observed in some junctions. The observation was largely without explanation until vesicles were shown by Peracchia using transmission electron microscopy (TEM) thin sections to be systematically associated with gap junction plaques.[134] Peracchia's study was probably also the first study to describe paired connexon structures, which he called a globule. Studies showing vesicles associated with gap junctions and proposing the vesicle contents may move across the junction plaques between two cells were rare, as most studies focused on connexons rather than vesicles. A later study using a combination of microscopy techniques confirmed the early evidence of a probable function for gap junctions in intercellular vesicle transfer. Areas of vesicle transfer were associated with connexin free islands within gap junction plaques.[114] Connexin 43 has been shown to be necessary for the transfer of whole mitochondrias to neighboring cells, though whether the mitochondria is transferred directly through the membrane or within a vesicle has not been determined [135]
Electrical and chemical synapses
Because of the widespread occurrence of gap junctions in cell types other than nerve cells, the term gap junction became more generally used than terms such as electrical synapse or nexus. Another dimension in the relationship between nerve cells and gap junctions was revealed by studying chemical synapse formation and gap junction presence. By tracing nerve development in leeches with gap junction expression suppressed it was shown that the bidirectional gap junction (electrical nerve synapse) needs to form between two cells before they can grow to form a unidirectional chemical nerve synapse.[136] The chemical nerve synapse is the synapse most often truncated to the more ambiguous term nerve synapse.
Composition
Connexins
The purification[137][138] of the intercellular gap junction plaques enriched in the channel forming protein (connexin) showed a protein forming hexagonal arrays in x-ray diffraction. Because of this, the systematic study and identification of the predominant gap junction protein[139] became possible. Refined ultrastructural studies by TEM[140][141] showed protein occurred in a complementary fashion in both cells participating in a gap junction plaque. The gap junction plaque is a relatively large area of membrane observed in TEM thin section and freeze fracture (FF) seen filled with transmembrane proteins in both tissues and more gently treated gap junction preparations. With the apparent ability for one protein alone to enable intercellular communication seen in gap junctions[142] the term gap junction tended to become synonymous with a group of assembled connexins though this was not shown in vivo. Biochemical analysis of gap junction isolated from various tissues demonstrated a family of connexins.[143][144][145]
The ultrastructure and biochemistry of isolated gap junctions already referenced had indicated the connexins preferentially group in gap junction plaques or domains and connexins were the best characterized constituent. It has been noted that the organisation of proteins into arrays with a gap junction plaque may be significant.[56][146] It is likely this early work was already reflecting the presence of more than just connexins in gap junctions. Combining the emerging fields of freeze-fracture to see inside membranes and immunocytochemistry to label cell components (Freeze-fracture replica immunolabelling or FRIL and thin section immunolabelling) showed gap junction plaques in vivo contained the connexin protein.[147][113] Later studies using immunofluorescence microscopy of larger areas of tissue clarified diversity in earlier results. Gap junction plaques were confirmed to have variable composition being home to connexon and non-connexin proteins as well making the modern usage of the terms "gap junction" and "gap junction plaque" non-interchangeable.[8] To summarize, in early literature the term "gap junction" referred to the regular gap between membranes in vertebrates and non-vertebrates apparently bridged by "globules". The junction correlated with the cell's ability to directly couple with its neighbors through pores in their membranes. Then for a while gap junctions were only referring to a structure that contains connexins and nothing more was thought to be involved. Later, the gap junction "plaque" was also found to contain other molecules that helped define it and make it function.
The "plaque" or "formation plaque"
File:Connexin43-Modulates-Cell-Polarity-and-Directional-Cell-Migration-by-Regulating-Microtubule-Dynamics-pone.0026379.s004.ogv Early descriptions of gap junctions, connexons or innexons did not refer to them as such; many other terms were used. It is likely that synaptic disks[148] were an accurate reference to gap junction plaques. While the detailed structure and function of the connexon was described in a limited way at the time the gross disk structure was relatively large and easily seen by various TEM techniques. Disks allowed researchers using TEM to easily locate the connexons contained within the disk like patches in vivo and in vitro. The disk or plaque appeared to have structural properties different from those imparted by the connexons/innexons alone.[64] It was thought that if the area of membrane in the plaque transmitted signals, the area of membrane would have to be sealed in some way to prevent leakage.[149] Later studies showed gap junction plaques are home to non-connexin proteins, making the modern usage of the terms "gap junction" and "gap junction plaque" non-interchangeable as the area of the gap junction plaque may contain proteins other than connexins.[8][114] Just as connexins do not always occupy the entire area of the plaque, the other components described in the literature may be only long-term or short-term residents.[150][32][151]
Studies allowing views inside the plane of the membrane of gap junctions during formation indicated that a "formation plaque" formed between two cells prior to the connexins moving in. They were particle free areas—when observed by TEM FF, indicated very small or no transmembrane proteins were likely present. Little is known about what structures make up the formation plaque or how the formation plaque's structure changes when connexins and other components move in and out. One of the earlier studies of the formation of small gap junctions describes rows of particles and particle free halos.[152] With larger gap junctions they were described as formation plaques with connexins moving into them. The particulate gap junctions were thought to form 4–6 hours after the formation plaques appeared.[153] How the connexins may be transported to the plaques using tubulin is becoming clearer.[85][154]
The formation of plaque and the non-connexin part of the classical gap junction plaque have been difficult for early researchers to analyse. It appears in TEM FF and thin section to be a lipid membrane domain that can somehow form a comparatively rigid barrier to other lipids and proteins. There has been indirect evidence for certain lipids being preferentially involved with the formation plaque, however this cannot be considered definitive.[155][156] It is difficult to envisage breaking up the membrane to analyse membrane plaques without affecting their composition. By study of connexins still in membranes lipids associated with the connexins have been studied.[157] It was found that specific connexins tended to associate preferentially with specific phospholipids. As formation plaques precede connexins these results still give no certainty as to what is unique about the composition of plaques themselves. Other findings show connexins associate with protein scaffolds used in another junction, the zonula occludens ZO-1.[158] While this helps us understand how connexins may be moved into a gap junction formation plaque, the composition of the plaque itself is still somewhat sketchy. Some headway on the in vivo composition of the gap junction plaque is being made using TEM FRIL.[150][158]
See also
- Gap junction modulation
- Gap junction protein
- Innexin
- Vinnexin
- Intercalated disc
- Ion channel
- Junctional complex
- Tight junction
References
- ↑ Talukdar, S; Emdad, L; Das, SK; Fisher, PB (2 January 2022). "GAP junctions: multifaceted regulators of neuronal differentiation.". Tissue Barriers 10 (1). doi:10.1080/21688370.2021.1982349. PMID 34651545.
- ↑ "Junctions between intimately apposed cell membranes in the vertebrate brain". J. Cell Biol. 40 (3): 648–677. March 1969. doi:10.1083/jcb.40.3.648. PMID 5765759.
- ↑ Revel, J. P.; Karnovsky, M. J. (1 June 1967). "Hexagonal Array of Subunits in Intercellular Junctions of the Mouse Heart and Liver". Journal of Cell Biology 33 (3): C7–12. doi:10.1083/jcb.33.3.C7. PMID 6036535.
- ↑ Hervé, Jean-Claude; Bourmeyster, Nicolas; Sarrouilhe, Denis; Duffy, Heather S. (May 2007). "Gap junctional complexes: From partners to functions". Progress in Biophysics and Molecular Biology 94 (1–2): 29–65. doi:10.1016/j.pbiomolbio.2007.03.010. PMID 17507078.
- ↑ 5.0 5.1 Gilleron, Jérome; Carette, Diane; Fiorini, Céline; Benkdane, Merieme; Segretain, Dominique; Pointis, Georges (March 2009). "Connexin 43 gap junction plaque endocytosis implies molecular remodelling of ZO-1 and c-Src partners". Communicative & Integrative Biology 2 (2): 104–106. doi:10.4161/cib.7626. PMID 19704902.
- ↑ 6.0 6.1 6.2 Ivanovic, Ena; Kucera, Jan P. (November 2021). "Localization of Na + channel clusters in narrowed perinexi of gap junctions enhances cardiac impulse transmission via ephaptic coupling: a model study". The Journal of Physiology 599 (21): 4779–4811. doi:10.1113/JP282105. PMID 34533834.
- ↑ Yu, Xun Sean; Yin, Xinye; Lafer, Eileen M.; Jiang, Jean X. (June 2005). "Developmental Regulation of the Direct Interaction between the Intracellular Loop of Connexin 45.6 and the C Terminus of Major Intrinsic Protein (Aquaporin-0)". Journal of Biological Chemistry 280 (23): 22081–22090. doi:10.1074/jbc.M414377200. PMID 15802270.
- ↑ 8.0 8.1 8.2 Gruijters, WTM (1989). "A non-connexon protein (MIP) is involved in eye lens gap-junction formation". Journal of Cell Science 93 (3): 509–513. doi:10.1242/jcs.93.3.509. PMID 2691517. http://jcs.biologists.org/cgi/content/abstract/93/3/509.
- ↑ Phelan, Pauline; Stebbings, Lucy A.; Baines, Richard A.; Bacon, Jonathan P.; Davies, Jane A.; Ford, Chris (January 1998). "Drosophila Shaking-B protein forms gap junctions in paired Xenopus oocytes". Nature 391 (6663): 181–184. doi:10.1038/34426. PMID 9428764. Bibcode: 1998Natur.391..181P.
- ↑ Phelan, Pauline; Bacon, Jonathan P.; A. Davies, Jane; Stebbings, Lucy A.; Todman, Martin G. (September 1998). "Innexins: a family of invertebrate gap-junction proteins". Trends in Genetics 14 (9): 348–349. doi:10.1016/s0168-9525(98)01547-9. PMID 9769729.
- ↑ Ortiz, Jennifer; Bobkov, Yuriy V; DeBiasse, Melissa B; Mitchell, Dorothy G; Edgar, Allison; Martindale, Mark Q; Moss, Anthony G; Babonis, Leslie S et al. (3 February 2023). "Independent Innexin Radiation Shaped Signaling in Ctenophores". Molecular Biology and Evolution 40 (2). doi:10.1093/molbev/msad025. PMID 36740225.
- ↑ 12.0 12.1 Principles of neural science (5th ed.). New York: McGraw-Hill medical. 2013. ISBN 978-0-07-139011-8.
- ↑ 13.0 13.1 Purves, Dale, ed (2001). Neuroscience (2nd ed.). Sunderland, Mass: Sinauer Associates. ISBN 978-0-87893-742-4.
- ↑ Furshpan, E. J.; Potter, D. D. (August 1957). "Mechanism of Nerve-Impulse Transmission at a Crayfish Synapse". Nature 180 (4581): 342–343. doi:10.1038/180342a0. PMID 13464833. Bibcode: 1957Natur.180..342F.
- ↑ Lampe, Paul D.; Lau, Alan F. (2004). "The effects of connexin phosphorylation on gap junctional communication". The International Journal of Biochemistry & Cell Biology 36 (7): 1171–86. doi:10.1016/S1357-2725(03)00264-4. PMID 15109565.
- ↑ Lampe, Paul D.; Lau, Alan F. (2000). "Regulation of gap junctions by phosphorylation of connexins". Archives of Biochemistry and Biophysics 384 (2): 205–15. doi:10.1006/abbi.2000.2131. PMID 11368307.
- ↑ Scemes, Eliana; Spray, David C.; Meda, Paolo (April 2009). "Connexins, pannexins, innexins: novel roles of "hemi-channels"". Pflügers Archiv: European Journal of Physiology 457 (6): 1207–1226. doi:10.1007/s00424-008-0591-5. PMID 18853183.
- ↑ Martinez-Banaclocha, Marcos (13 February 2020). "Astroglial Isopotentiality and Calcium-Associated Biomagnetic Field Effects on Cortical Neuronal Coupling". Cells 9 (2): 439. doi:10.3390/cells9020439. PMID 32069981.
- ↑ Parker, David (22 December 2022). "Neurobiological reduction: From cellular explanations of behavior to interventions". Frontiers in Psychology 13. doi:10.3389/fpsyg.2022.987101. PMID 36619115.
- ↑ Peracchia, Camillo (1 April 1973). "Low Resistance Junctions in Crayfish". Journal of Cell Biology 57 (1): 66–76. doi:10.1083/jcb.57.1.66. PMID 4120611.
- ↑ 21.0 21.1 Maeda, Shoji; Nakagawa, So; Suga, Michihiro; Yamashita, Eiki; Oshima, Atsunori; Fujiyoshi, Yoshinori; Tsukihara, Tomitake (2009). "Structure of the connexin 26 gap junction channel at 3.5 A resolution". Nature 458 (7238): 597–602. doi:10.1038/nature07869. PMID 19340074. Bibcode: 2009Natur.458..597M.
- ↑ Perkins, Guy A.; Goodenough, Daniel A.; Sosinsky, Gina E. (1998). "Formation of the gap junction intercellular channel requires a 30 degree rotation for interdigitating two apposing connexons". Journal of Molecular Biology 277 (2): 171–7. doi:10.1006/jmbi.1997.1580. PMID 9514740.
- ↑ The C. elegans Sequencing Consortium (Dec 11, 1998). "Genome sequence of the nematode C. elegans: a platform for investigating biology". Science 282 (5396): 2012–2018. doi:10.1126/science.282.5396.2012. PMID 9851916. Bibcode: 1998Sci...282.2012..
- ↑ Ganfornina, MD; Sánchez, D; Herrera, M; Bastiani, MJ (1999). "Developmental expression and molecular characterization of two gap junction channel proteins expressed during embryogenesis in the grasshopper Schistocerca americana". Developmental Genetics 24 (1–2): 137–150. doi:10.1002/(SICI)1520-6408(1999)24:1/2<137::AID-DVG13>3.0.CO;2-7. PMID 10079517.
- ↑ Starich, T. A. (1996). "eat-5 and unc-7 represent a multigene family in Caenorhabditis elegans involved in cell-cell coupling". J. Cell Biol. 134 (2): 537–548. doi:10.1083/jcb.134.2.537. PMID 8707836.
- ↑ Simonsen, Karina T.; Moerman, Donald G.; Naus, Christian C. (2014). "Gap junctions in C. elegans". Frontiers in Physiology 5: 40. doi:10.3389/fphys.2014.00040. PMID 24575048.
- ↑ Barbe, M. T. (1 April 2006). "Cell-Cell Communication Beyond Connexins: The Pannexin Channels". Physiology 21 (2): 103–114. doi:10.1152/physiol.00048.2005. PMID 16565476.
- ↑ Panchina, Yuri; Kelmanson, Ilya; Matz, Mikhail; Lukyanov, Konstantin; Usman, Natalia; Lukyanov, Sergey (June 2000). "A ubiquitous family of putative gap junction molecules". Current Biology 10 (13): R473–R474. doi:10.1016/S0960-9822(00)00576-5. PMID 10898987. Bibcode: 2000CBio...10.R473P.
- ↑ Lohman, Alexander W.; Isakson, Brant E. (2014). "Differentiating connexin hemichannels and pannexin channels in cellular ATP release". FEBS Letters 588 (8): 1379–1388. doi:10.1016/j.febslet.2014.02.004. PMID 24548565. Bibcode: 2014FEBSL.588.1379L.
- ↑ "Pannexin channels are not gap junction hemichannels". Channels 5 (3): 193–197. 2011-05-01. doi:10.4161/chan.5.3.15765. PMID 21532340.
- ↑ Slivko-Koltchik, Georgy A. (2019-02-26). "Are there gap junctions without connexins or pannexins?". BMC Ecol. Evol. 19 (Suppl 1). doi:10.1186/s12862-019-1369-4. PMID 30813901. Bibcode: 2019BMCEE..19S..46S.
- ↑ 32.0 32.1 Hervé, JC; Bourmeyster, N; Sarrouilhe, D; Duffy, HS (May 2007). "Gap junctional complexes: from partners to functions.". Prog Biophys Mol Biol 94 (1–2): 29–65. doi:10.1016/j.pbiomolbio.2007.03.010. PMID 17507078.
- ↑ Hsieh, CL; Kumar, NM; Gilula, NB; Francke, U (Mar 1991). "Distribution of genes for gap junction membrane channel proteins on human and mouse chromosomes.". Somatic Cell and Molecular Genetics 17 (2): 191–200. doi:10.1007/bf01232976. PMID 1849321.
- ↑ Kumar, NM; Gilula, NB (Feb 1992). "Molecular biology and genetics of gap junction channels.". Seminars in Cell Biology 3 (1): 3–16. doi:10.1016/s1043-4682(10)80003-0. PMID 1320430.
- ↑ Kren, BT; Kumar, NM; Wang, SQ; Gilula, NB; Steer, CJ (Nov 1993). "Differential regulation of multiple gap junction transcripts and proteins during rat liver regeneration.". The Journal of Cell Biology 123 (3): 707–18. doi:10.1083/jcb.123.3.707. PMID 8227133.
- ↑ Oshima, A; Tani, K; Fujiyoshi, Y (1 Dec 2016). "Atomic structure of the innexin-6 gap junction channel determined by cryo-EM.". Nat. Commun. 7. doi:10.1038/ncomms13681. PMID 27905396. Bibcode: 2016NatCo...713681O.
- ↑ 37.0 37.1 Lauf, Undine; Giepmans, Ben N. G.; Lopez, Patricia; Braconnot, Sébastien; Chen, Shu-Chih; Falk, Matthias M. (6 August 2002). "Dynamic trafficking and delivery of connexons to the plasma membrane and accretion to gap junctions in living cells". Proceedings of the National Academy of Sciences 99 (16): 10446–10451. doi:10.1073/pnas.162055899. PMID 12149451. Bibcode: 2002PNAS...9910446L.
- ↑ Chang, Qing; Tang, Wenxue; Ahmad, Shoeb; Zhou, Binfei; Lin, Xi (2008). Schiffmann, Raphael. ed. "Gap junction mediated intercellular metabolite transfer in the cochlea is compromised in connexin30 null mice". PLOS ONE 3 (12): e4088. doi:10.1371/journal.pone.0004088. PMID 19116647. Bibcode: 2008PLoSO...3.4088C.
- ↑ Alberts, Bruce (2002). Molecular biology of the cell (4th ed.). New York: Garland Science. ISBN 978-0-8153-3218-3.
- ↑ "Exchange of conductance and gating properties between gap junction hemichannels". FEBS Lett 451 (2): 113–117. 1999. doi:10.1016/S0014-5793(99)00558-X. PMID 10371149. Bibcode: 1999FEBSL.451..113H.
- ↑ "Permeability of membrane junctions". Ann. N. Y. Acad. Sci. 137 (2): 441–472. July 1966. doi:10.1111/j.1749-6632.1966.tb50175.x. PMID 5229810. Bibcode: 1966NYASA.137..441L.
- ↑ Khan, Ali K.; Jagielnicki, Maciej; Bennett, Brad.C.; Purdy, Michael D.; Yeager, Mark (September 2021). "Cryo-EM structure of an open conformation of a gap junction hemichannel in lipid bilayer nanodiscs". Structure 29 (9): 1040–1047.e3. doi:10.1016/j.str.2021.05.010. PMID 34129834.
- ↑ Goodenough, Daniel A.; Paul, David L. (April 2003). "Beyond the gap: functions of unpaired connexon channels". Nature Reviews Molecular Cell Biology 4 (4): 285–295. doi:10.1038/nrm1072. PMID 12671651.
- ↑ Laird, Dale W.; Lampe, Paul D. (December 2018). "Therapeutic strategies targeting connexins". Nature Reviews Drug Discovery 17 (12): 905–921. doi:10.1038/nrd.2018.138. PMID 30310236.
- ↑ 45.0 45.1 45.2 Van Campenhout, Raf; Gomes, Ana Rita; De Groof, Timo W.M.; Muyldermans, Serge; Devoogdt, Nick; Vinken, Mathieu (28 March 2021). "Mechanisms Underlying Connexin Hemichannel Activation in Disease". International Journal of Molecular Sciences 22 (7): 3503. doi:10.3390/ijms22073503. PMID 33800706.
- ↑ "Cell contacts in human islets of Langerhans". J. Clin. Endocrinol. Metab. 41 (5): 841–4. November 1975. doi:10.1210/jcem-41-5-841. PMID 1102552. http://jcem.endojournals.org/cgi/pmidlookup?view=long&pmid=1102552.
- ↑ Garfield, RE; Sims, SM; Kannan, MS; Daniel, EE (November 1978). "Possible role of gap junctions in activation of myometrium during parturition". Am. J. Physiol. 235 (5): C168–79. doi:10.1152/ajpcell.1978.235.5.C168. PMID 727239.
- ↑ Goodenough, DA (November 1979). "Lens gap junctions: a structural hypothesis for nonregulated low-resistance intercellular pathways". Invest. Ophthalmol. Vis. Sci. 18 (11): 1104–22. PMID 511455.
- ↑ "Variations in tight and gap junctions in mammalian tissues". J. Cell Biol. 53 (3): 758–76. June 1972. doi:10.1083/jcb.53.3.758. PMID 4337577.
- ↑ "Intercellular junctional complexes of the rat seminiferous tubules: a freeze-fracture study". Anat. Rec. 189 (2): 211–31. October 1977. doi:10.1002/ar.1091890208. PMID 911045.
- ↑ Kreutziger GO (September 1976). "Lateral membrane morphology and gap junction structure in rabbit corneal endothelium". Exp. Eye Res. 23 (3): 285–93. doi:10.1016/0014-4835(76)90129-9. PMID 976372.
- ↑ Albertini, DF; Anderson, E. (Oct 1974). "The appearance and structure of intercellular connections during the ontogeny of the rabbit ovarian follicle with particular reference to gap junctions". J Cell Biol 63 (1): 234–50. doi:10.1083/jcb.63.1.234. PMID 4417791.
- ↑ Prutkin L (February 1975). "Mucous metaplasia and gap junctions in the vitamin A acid-treated skin tumor, keratoacanthoma". Cancer Res. 35 (2): 364–9. PMID 1109802. http://cancerres.aacrjournals.org/cgi/pmidlookup?view=long&pmid=1109802.
- ↑ Raviola, E; Gilula, NB (Jun 1973). "Gap junctions between photoreceptor cells in the vertebrate retina". Proc Natl Acad Sci U S A 70 (6): 1677–81. doi:10.1073/pnas.70.6.1677. PMID 4198274. Bibcode: 1973PNAS...70.1677R.
- ↑ Bellairs, R; Breathnach, AS; Gross, M (Sep 1975). "Freeze-fracture replication of junctional complexes in unincubated and incubated chick embryos". Cell Tissue Res. 162 (2): 235–52. doi:10.1007/BF00209209. PMID 1237352.
- ↑ 56.0 56.1 J. Cell Biol. 1974 Jul;62(1) 32-47.Assembly of gap junctions during amphibian neurulation. Decker RS, Friend DS.
- ↑ "Differentiation of the junctional complex of surface cells in the developing Fundulus blastoderm". J. Cell Biol. 48 (3): 455–72. March 1971. doi:10.1083/jcb.48.3.455. PMID 5545331.
- ↑ 58.0 58.1 Robertson, JD (February 1953). "Ultrastructure of two invertebrate synapses". Proceedings of the Society for Experimental Biology and Medicine 82 (2): 219–23. doi:10.3181/00379727-82-20071. PMID 13037850.
- ↑ Shibata, Y; Yamamoto, T (March 1977). "Gap junctions in the cardiac muscle cells of the lamprey". Cell Tissue Res. 178 (4): 477–82. doi:10.1007/BF00219569. PMID 870202.
- ↑ Lorber, V; Rayns, DG (April 1977). "Fine structure of the gap junction in the tunicate heart". Cell Tissue Res. 179 (2): 169–75. doi:10.1007/BF00219794. PMID 858161.
- ↑ "Gap junctions between the supporting cells in some acoustico-vestibular receptors". J. Neurocytol. 6 (1): 1–12. February 1977. doi:10.1007/BF01175410. PMID 839246.
- ↑ Hudspeth, AJ; Revel, JP (Jul 1971). "Coexistence of gap and septate junctions in an invertebrate epithelium". J. Cell Biol. 50 (1): 92–101. doi:10.1083/jcb.50.1.92. PMID 5563454.
- ↑ Boucaud-Camou, Eve (1980). "Junctional structures in digestive epithelia of a cephalopod". Tissue Cell 12 (2): 395–404. doi:10.1016/0040-8166(80)90013-0. PMID 7414602.
- ↑ 64.0 64.1 Hand, AR; Gobel, S (February 1972). "The structural organization of the septate and gap junctions of Hydra". J. Cell Biol. 52 (2): 397–408. doi:10.1083/jcb.52.2.397. PMID 4109925.
- ↑ Baerwald RJ (1975). "Inverted gap and other cell junctions in cockroach hemocyte capsules: a thin section and freeze-fracture study". Tissue Cell 7 (3): 575–85. doi:10.1016/0040-8166(75)90027-0. PMID 1179417.
- ↑ "Gap junction formation between reaggregated Novikoff hepatoma cells". Proc. Natl. Acad. Sci. U.S.A. 71 (11): 4536–40. November 1974. doi:10.1073/pnas.71.11.4536. PMID 4373716. Bibcode: 1974PNAS...71.4536J.
- ↑ Knudsen, KA; Horwitz, AF (1978). "Toward a mechanism of myoblast fusion". Prog Clin Biol Res 23: 563–8. PMID 96453.
- ↑ "The incidence and size of gap junctions between the bone cells in rat calvaria". Anat. Embryol. 187 (4): 343–52. April 1993. doi:10.1007/BF00185892. PMID 8390141.
- ↑ Sperelakis, Nicholas; Ramasamy, Lakshminarayanan (2005). "Gap-junction channels inhibit transverse propagation in cardiac muscle". Biomed Eng Online 4 (1): 7. doi:10.1186/1475-925X-4-7. PMID 15679888.
- ↑ "Intercellular communication and tissue growth: IX. Junctional membrane structure of hybrids between communication-competent and communication-incompetent cells". J. Membr. Biol. 34 (1): 39–54. June 1977. doi:10.1007/BF01870292. PMID 561191.
- ↑ "Comparison of contact-mediated communication in normal and transformed human cells in culture". Proc. Natl. Acad. Sci. U.S.A. 74 (10): 4476–80. October 1977. doi:10.1073/pnas.74.10.4476. PMID 270694. Bibcode: 1977PNAS...74.4476C.
- ↑ Habermann, H; Chang, WY; Birch, L; Mehta, P; Prins, GS (January 2001). "Developmental exposure to estrogens alters epithelial cell adhesion and gap junction proteins in the adult rat prostate". Endocrinology 142 (1): 359–69. doi:10.1210/endo.142.1.7893. PMID 11145599.
- ↑ Kelley, Robert O.; Vogel, Kathryn G.; Crissman, Harry A.; Lujan, Christopher J.; Skipper, Betty E. (March 1979). "Development of the aging cell surface. Reduction of gap junction-mediated metabolic cooperation with progressive subcultivation of human embryo fibroblasts (IMR-90)". Exp. Cell Res. 119 (1): 127–43. doi:10.1016/0014-4827(79)90342-2. PMID 761600.
- ↑ Phelan, Pauline (June 2005). "Innexins: members of an evolutionarily conserved family of gap-junction proteins". Biochimica et Biophysica Acta (BBA) - Biomembranes 1711 (2): 225–245. doi:10.1016/j.bbamem.2004.10.004. PMID 15921654.
- ↑ Hervé, Jean-Claude; Phelan, Pauline; Bruzzone, Roberto; White, Thomas W. (December 2005). "Connexins, innexins and pannexins: Bridging the communication gap". Biochimica et Biophysica Acta (BBA) - Biomembranes 1719 (1–2): 3–5. doi:10.1016/j.bbamem.2005.11.013. PMID 16359939.
- ↑ Güiza, Juan; García, Aníbal; Arriagada, Javiera; Gutiérrez, Camila; González, Jorge; Márquez-Miranda, Valeria; Alegría-Arcos, Melissa; Duarte, Yorley et al. (February 2022). "Unnexins: Homologs of innexin proteins in Trypanosomatidae parasites". Journal of Cellular Physiology 237 (2): 1547–1560. doi:10.1002/jcp.30626. PMID 34779505.
- ↑ Turnbull, Matthew W.; Volkoff, Anne-Nathalie; Webb, Bruce A.; Phelan, Pauline (July 2005). "Functional gap junction genes are encoded by insect viruses". Current Biology 15 (13): R491–R492. doi:10.1016/j.cub.2005.06.052. PMID 16005277. Bibcode: 2005CBio...15.R491T.
- ↑ Moroz, Leonid L.; Romanova, Daria Y. (23 December 2022). "Alternative neural systems: What is a neuron? (Ctenophores, sponges and placozoans)". Frontiers in Cell and Developmental Biology 10. doi:10.3389/fcell.2022.1071961. PMID 36619868.
- ↑ Bergquist, P.R.; Green, C.R. (1977). "An ultrastructural study of settlement and metamorphosis in sponge larvae". Cah. Biol. Mar. 18: 289–302.
- ↑ Green, C.R.; Bergquist, P.R. (1979). "Cell membrane specializations in the Porifera". Coll Int Cent Natn Res Scient 291: 153–158.
- ↑ Warner, Anne E.; Guthrie, Sarah C.; Gilula, Norton B. (1984). "Antibodies to gap-junctional protein selectively disrupt junctional communication in the early amphibian embryo". Nature 311 (5982): 127–31. doi:10.1038/311127a0. PMID 6088995. Bibcode: 1984Natur.311..127W.
- ↑ Warner, AE (2007). "The Use of Antibodies to Gap Junction Protein to Explore the Role of Gap Junctional Communication During Development". Ciba Foundation Symposium 125 - Junctional Complexes of Epithelial Cells. Novartis Foundation Symposia. 125. pp. 154–67. doi:10.1002/9780470513408.ch10. ISBN 978-0-470-51340-8.
- ↑ Bastide, B; Jarry-Guichard, T; Briand, JP; Délèze, J; Gros, D (April 1996). "Effect of antipeptide antibodies directed against three domains of connexin43 on the gap junctional permeability of cultured heart cells". J. Membr. Biol. 150 (3): 243–53. doi:10.1007/s002329900048. PMID 8661989.
- ↑ Hofer, A; Dermietzel, R (September 1998). "Visualization and functional blocking of gap junction hemichannels (connexons) with antibodies against external loop domains in astrocytes". Glia 24 (1): 141–54. doi:10.1002/(SICI)1098-1136(199809)24:1<141::AID-GLIA13>3.0.CO;2-R. PMID 9700496.
- ↑ 85.0 85.1 85.2 Brandner, Johanna M, ed (2011). "Connexin43 modulates cell polarity and directional cell migration by regulating microtubule dynamics". PLOS ONE 6 (10). doi:10.1371/journal.pone.0026379. PMID 22022608. Bibcode: 2011PLoSO...626379F.
- ↑ Levin, Michael; Mercola, Mark (November 1998). "Gap junctions are involved in the early generation of left-right asymmetry". Dev. Biol. 203 (1): 90–105. doi:10.1006/dbio.1998.9024. PMID 9806775.
- ↑ Levin, M; Mercola, M (November 1999). "Gap junction-mediated transfer of left-right patterning signals in the early chick blastoderm is upstream of Shh asymmetry in the node". Development 126 (21): 4703–14. doi:10.1242/dev.126.21.4703. PMID 10518488.
- ↑ Bani-Yaghoub, Mahmud; Underhill, T. Michael; Naus, Christian C.G. (1999). "Gap junction blockage interferes with neuronal and astroglial differentiation of mouse P19 embryonal carcinoma cells". Dev. Genet. 24 (1–2): 69–81. doi:10.1002/(SICI)1520-6408(1999)24:1/2<69::AID-DVG8>3.0.CO;2-M. PMID 10079512.
- ↑ Bani-Yaghoub, Mahmud; Bechberger, John F.; Underhill, T. Michael; Naus, Christian C. G. (March 1999). "The effects of gap junction blockage on neuronal differentiation of human NTera2/clone D1 cells". Exp. Neurol. 156 (1): 16–32. doi:10.1006/exnr.1998.6950. PMID 10192774.
- ↑ Donahue, HJ; Li, Z; Zhou, Z; Yellowley, CE (February 2000). "Differentiation of human fetal osteoblastic cells and gap junctional intercellular communication". Am. J. Physiol., Cell Physiol. 278 (2): C315–22. doi:10.1152/ajpcell.2000.278.2.C315. PMID 10666026.
- ↑ Cronier, L; Frendo, JL; Defamie, N; Pidoux, G; Bertin, G; Guibourdenche, J; Pointis, G; Malassine, A (November 2003). "Requirement of gap junctional intercellular communication for human villous trophoblast differentiation". Biol. Reprod. 69 (5): 1472–80. doi:10.1095/biolreprod.103.016360. PMID 12826585.
- ↑ El-Sabban, ME; Sfeir, AJ; Daher, MH; Kalaany, NY; Bassam, RA; Talhouk, RS (September 2003). "ECM-induced gap junctional communication enhances mammary epithelial cell differentiation". J. Cell Sci. 116 (Pt 17): 3531–41. doi:10.1242/jcs.00656. PMID 12893812.
- ↑ Chaytor, AT; Martin, PE; Evans, WH; Randall, MD; Griffith, TM (October 1999). "The endothelial component of cannabinoid-induced relaxation in rabbit mesenteric artery depends on gap junctional communication". J. Physiol. 520 (2): 539–50. doi:10.1111/j.1469-7793.1999.00539.x. PMID 10523421.
- ↑ Srinivas, M; Hopperstad, MG; Spray, DC (September 2001). "Quinine blocks specific gap junction channel subtypes". Proc. Natl. Acad. Sci. U.S.A. 98 (19): 10942–7. doi:10.1073/pnas.191206198. PMID 11535816. Bibcode: 2001PNAS...9810942S.
- ↑ Li Bi, Wan; Parysek, Linda M.; Warnick, Ronald; Stambrook, Peter J. (December 1993). "In vitro evidence that metabolic cooperation is responsible for the bystander effect observed with HSV tk retroviral gene therapy". Hum. Gene Ther. 4 (6): 725–31. doi:10.1089/hum.1993.4.6-725. PMID 8186287.
- ↑ Little, JB; Azzam, EI; De Toledo, SM; Nagasawa, H (2002). "Bystander effects: intercellular transmission of radiation damage signals". Radiat Prot Dosimetry 99 (1–4): 159–62. doi:10.1093/oxfordjournals.rpd.a006751. PMID 12194273.
- ↑ Zhou, H; Randers-Pehrson, G; Suzuki, M; Waldren, CA; Hei, TK (2002). "Genotoxic damage in non-irradiated cells: contribution from the bystander effect". Radiat Prot Dosimetry 99 (1–4): 227–32. doi:10.1093/oxfordjournals.rpd.a006769. PMID 12194291.
- ↑ Lorimore, SA; Wright, EG (January 2003). "Radiation-induced genomic instability and bystander effects: related inflammatory-type responses to radiation-induced stress and injury? A review". Int. J. Radiat. Biol. 79 (1): 15–25. doi:10.1080/0955300021000045664. PMID 12556327.
- ↑ Ehrlich, HP; Diez, T (2003). "Role for gap junctional intercellular communications in wound repair". Wound Repair Regen 11 (6): 481–9. doi:10.1046/j.1524-475X.2003.11616.x. PMID 14617290.
- ↑ Coutinho, P.; Qiu, C.; Frank, S.; Wang, C.M.; Brown, T.; Green, C.R.; Becker, D.L. (July 2005). "Limiting burn extension by transient inhibition of Connexin43 expression at the site of injury". Br J Plast Surg 58 (5): 658–67. doi:10.1016/j.bjps.2004.12.022. PMID 15927148.
- ↑ Wang, C. M.; Lincoln, J.; Cook, J. E.; Becker, D. L. (November 2007). "Abnormal connexin expression underlies delayed wound healing in diabetic skin". Diabetes 56 (11): 2809–17. doi:10.2337/db07-0613. PMID 17717278.
- ↑ Rivera, EM; Vargas, M; Ricks-Williamson, L (1997). "Considerations for the aesthetic restoration of endodontically treated anterior teeth following intracoronal bleaching". Pract Periodontics Aesthet Dent 9 (1): 117–28. PMID 9550065.
- ↑ Mugisho, Odunayo O.; Aryal, Jyoti; Shorne, Avik; Lyon, Heather; Acosta, Monica L.; Green, Colin R.; Rupenthal, Ilva D. (15 February 2023). "Orally Delivered Connexin43 Hemichannel Blocker, Tonabersat, Inhibits Vascular Breakdown and Inflammasome Activation in a Mouse Model of Diabetic Retinopathy". International Journal of Molecular Sciences 24 (4): 3876. doi:10.3390/ijms24043876. PMID 36835288.
- ↑ Cusato, K; Bosco, A; Rozental, R; Guimarães, CA; Reese, BE; Linden, R; Spray, DC (July 2003). "Gap junctions mediate bystander cell death in developing retina". J. Neurosci. 23 (16): 6413–22. doi:10.1523/JNEUROSCI.23-16-06413.2003. PMID 12878681.
- ↑ Moyer, Kurtis E.; Saggers, Gregory C.; Ehrlich, H. Paul (2004). "Mast cells promote fibroblast populated collagen lattice contraction through gap junction intercellular communication". Wound Repair Regen 12 (3): 269–75. doi:10.1111/j.1067-1927.2004.012310.x. PMID 15225205.
- ↑ Djalilian, A. R.; McGaughey, D; Patel, S; Seo, EY; Yang, C; Cheng, J; Tomic, M; Sinha, S et al. (May 2006). "Connexin 26 regulates epidermal barrier and wound remodeling and promotes psoriasiform response". J. Clin. Invest. 116 (5): 1243–53. doi:10.1172/JCI27186. PMID 16628254.
- ↑ Zhang, Y.; Wang, H.; Kovacs, A.; Kanter, E. M.; Yamada, K. A. (February 2010). "Reduced expression of Cx43 attenuates ventricular remodeling after myocardial infarction via impaired TGF-beta signaling". Am. J. Physiol. Heart Circ. Physiol. 298 (2): H477–87. doi:10.1152/ajpheart.00806.2009. PMID 19966054.
- ↑ "TLR2 mediates gap junctional intercellular communication through connexin-43 in intestinal epithelial barrier injury". J. Biol. Chem. 284 (33): 22332–43. August 2009. doi:10.1074/jbc.M901619200. PMID 19528242.
- ↑ Xu, Ji; Nicholson, Bruce J. (January 2013). "The role of connexins in ear and skin physiology — Functional insights from disease-associated mutations". Biochimica et Biophysica Acta (BBA) - Biomembranes 1828 (1): 167–178. doi:10.1016/j.bbamem.2012.06.024. PMID 22796187.
- ↑ Srinivas, Miduturu; Verselis, Vytas K.; White, Thomas W. (1 January 2018). "Human diseases associated with connexin mutations". Biochimica et Biophysica Acta (BBA) - Biomembranes 1860 (1): 192–201. doi:10.1016/j.bbamem.2017.04.024. PMID 28457858.
- ↑ White, Thomas W.; Paul, David L. (1999). "Genetic diseases and gene knockouts reveal diverse connexin functions". Annual Review of Physiology 61 (1): 283–310. doi:10.1146/annurev.physiol.61.1.283. PMID 10099690.
- ↑ Ivanovic, Ena; Kucera, Jan P. (2 November 2022). "Tortuous Cardiac Intercalated Discs Modulate Ephaptic Coupling". Cells 11 (21): 3477. doi:10.3390/cells11213477. PMID 36359872.
- ↑ 113.0 113.1 Gruijters, W.T.; Kistler, J.; Bullivant, S. (1 October 1987). "Formation, distribution and dissociation of intercellular junctions in the lens". Journal of Cell Science 88 (3): 351–359. doi:10.1242/jcs.88.3.351. PMID 3448099.
- ↑ 114.0 114.1 114.2 Gruijters, W (2003). "Are gap junction membrane plaques implicated in intercellular vesicle transfer?". Cell Biology International 27 (9): 711–717. doi:10.1186/s12862-019-1369-4. PMID 30813901. Bibcode: 2019BMCEE..19S..46S.
- ↑ Connors; Long (2004). "Electrical synapses in the mammalian brain". Annu Rev Neurosci 27: 393–418. doi:10.1146/annurev.neuro.26.041002.131128. PMID 15217338. https://zenodo.org/record/894386.
- ↑ 116.0 116.1 Ammer, Georg; Vieira, Renee M; Fendl, Sandra; Borst, Alexander (2022). "Anatomical distribution and functional roles of electrical synapses in Drosophila". Current Biology (Elsevier) 32 (9): 2022–2036.e4. doi:10.1016/j.cub.2022.03.040. PMID 35385694. Bibcode: 2022CBio...32E2022A.
- ↑ Orthmann-Murphy, Jennifer L.; Abrams, Charles K.; Scherer, Steven S. (May 2008). "Gap Junctions Couple Astrocytes and Oligodendrocytes". Journal of Molecular Neuroscience 35 (1): 101–116. doi:10.1007/s12031-007-9027-5. PMID 18236012.
- ↑ Wu, Chia-Lin; Shih, Meng-Fu Maxwell; Lai, Jason Sih-Yu; Yang, Hsun-Ti; Turner, Glenn C; Chen, Linyi; Chiang, Ann-Shyn (2011). "Heterotypic gap junctions between two neurons in the Drosophila brain are critical for memory". Current Biology (Elsevier) 21 (10): 848–854. doi:10.1016/j.cub.2011.02.041. PMID 21530256. Bibcode: 2011CBio...21..848W.
- ↑ Liu, Qingqing; Yang, Xing; Tian, Jingsong; Gao, Zhongbao; Wang, Meng; Li, Yan; Guo, Aike (2016). "Gap junction networks in mushroom bodies participate in visual learning and memory in Drosophila". eLife (eLife Sciences Publications, Ltd) 5. doi:10.7554/eLife.13238. PMID 27218450. PMC 4909397. https://elifesciences.org/articles/13238.pdf.
- ↑ Pannasch, Ulrike; Vargová, Lydia; Reingruber, Jürgen; Ezan, Pascal; Holcman, David; Giaume, Christian; Syková, Eva; Rouach, Nathalie (2011-05-17). "Astroglial networks scale synaptic activity and plasticity" (in en). Proceedings of the National Academy of Sciences 108 (20): 8467–8472. doi:10.1073/pnas.1016650108. ISSN 0027-8424. PMID 21536893. Bibcode: 2011PNAS..108.8467P.
- ↑ Ghézali, Grégory; Dallérac, Glenn; Rouach, Nathalie (2016). "Perisynaptic astroglial processes: dynamic processors of neuronal information" (in en). Brain Structure and Function 221 (5): 2427–2442. doi:10.1007/s00429-015-1070-3. ISSN 1863-2653. PMID 26026482. http://link.springer.com/10.1007/s00429-015-1070-3.
- ↑ Hardy, Eléonore; Moulard, Julien; Walter, Augustin; Ezan, Pascal; Bemelmans, Alexis-Pierre; Mouthon, Franck; Charvériat, Mathieu; Rouach, Nathalie et al. (2023-04-11). Eroglu, Cagla. ed. "Upregulation of astroglial connexin 30 impairs hippocampal synaptic activity and recognition memory" (in en). PLOS Biology 21 (4). doi:10.1371/journal.pbio.3002075. ISSN 1545-7885. PMID 37040348. "Cx30 upregulation increases the connectivity of astroglial networks, it decreases spontaneous and evoked synaptic transmission. This effect results from a reduced neuronal excitability and translates into an alteration in the induction of synaptic plasticity and an in vivo impairment in learning processes. Altogether, these results suggest that astroglial networks have a physiologically optimized size to appropriately regulate neuronal functions.".
- ↑ Hardy, Eléonore; Cohen-Salmon, Martine; Rouach, Nathalie; Rancillac, Armelle (September 2021). "Astroglial Cx30 differentially impacts synaptic activity from hippocampal principal cells and interneurons" (in en). Glia 69 (9): 2178–2198. doi:10.1002/glia.24017. ISSN 0894-1491. PMID 33973274. https://onlinelibrary.wiley.com/doi/10.1002/glia.24017. "Cx30 differentially alters the electrophysiological and morphological properties of hippocampal cell populations. They modulates both excitatory and inhibitory inputs. Astrocytes, via Cx30, are thus active modulators of both excitatory and inhibitory synapses in the hippocampus.".
- ↑ Béla Völgyi; Stewart A. Bloomfield (February 2009). "The diverse functional roles and regulation of neuronal gap junctions in the retina". Nature Reviews Neuroscience 10 (7): 495–506. doi:10.1016/S0165-0173(99)00070-3. PMID 19491906.
- ↑ Garfield, RE; Sims, SM; Kannan, MS; Daniel, EE (November 1978). "Possible role of gap junctions in activation of myometrium during parturition". Am. J. Physiol. 235 (5): C168–79. doi:10.1152/ajpcell.1978.235.5.C168. PMID 727239. S2CID 31610495
- ↑ Boros-Rausch, A., Shynlova, O., & Lye, S. J. (2021). "A Broad-Spectrum Chemokine Inhibitor Blocks Inflammation-Induced Myometrial Myocyte-Macrophage Crosstalk and Myometrial Contraction". Cells. 11 (1): 128. doi: 10.3390/cells11010128 PMID 35011690
- ↑ Evans, W. Howard; De Vuyst, Elke; Leybaert, Luc (1 July 2006). "The gap junction cellular internet: connexin hemichannels enter the signalling limelight". Biochemical Journal 397 (1): 1–14. doi:10.1042/BJ20060175. PMID 16761954.
- ↑ Robertson, J. D. (1963). Locke, Michael. ed. Cellular membranes in development. New York: Academic Press. OCLC 261587041.
- ↑ Robertson (1981). "Membrane structure". The Journal of Cell Biology 91 (3): 189s–204s. doi:10.1083/jcb.91.3.189s. PMID 7033238.
- ↑ "Mechanism of Nerve-Impulse Transmission at a Crayfish Synapse". Nature 180 (4581): 342–343. 1957. doi:10.1038/180342a0. PMID 13464833. Bibcode: 1957Natur.180..342F.
- ↑ "Transmission at the giant motor synapses of the crayfish". The Journal of Physiology 145 (2): 289–325. 1959. doi:10.1113/jphysiol.1959.sp006143. PMID 13642302.
- ↑ Payton, B. W.; Bennett, M. V. L.; Pappas, G. D. (December 1969). "Permeability and structure of junctional membranes at an electrotonic synapse". Science 166 (3913): 1641–1643. doi:10.1126/science.166.3913.1641. PMID 5360587. Bibcode: 1969Sci...166.1641P.
- ↑ Chalcroft, J. P.; Bullivant, S (October 1970). "An interpretation of liver cell membrane and junction structure based on observation of freeze-fracture replicas of both sides of the fracture". J. Cell Biol. 47 (1): 49–60. doi:10.1083/jcb.47.1.49. PMID 4935338.
- ↑ Peracchia, C (April 1973). "Low resistance junctions in crayfish. II. Structural details and further evidence for intercellular channels by freeze-fracture and negative staining". J. Cell Biol. 57 (1): 54–65. doi:10.1083/jcb.57.1.54. PMID 4120610.
- ↑ Islam, M.Expression error: Unrecognized word "et". (2012). "Mitochondrial transfer from bone-marrow–derived stromal cells to pulmonary alveoli protects against acute lung injury.". Nat Med 18 (5): 759–765. doi:10.1038/nm.2736. PMID 22504485.
- ↑ "Gap junction expression is required for normal chemical synapse formation". J. Neurosci. 30 (45): 15277–85. November 2010. doi:10.1523/JNEUROSCI.2331-10.2010. PMID 21068332.
- ↑ Goodenough, D. A.; Stoeckenius, W. (1972). "The isolation of mouse hepatocyte gap junctions: Preliminary Chemical Characterization and X-Ray Diffraction". The Journal of Cell Biology 54 (3): 646–56. doi:10.1083/jcb.54.3.646. PMID 4339819.
- ↑ Goodenough, D. A. (1974). "Bulk isolation of mouse hepatocyte gap junctions: Characterization of the Principal Protein, Connexin". The Journal of Cell Biology 61 (2): 557–63. doi:10.1083/jcb.61.2.557. PMID 4363961.
- ↑ Kumar, N. M.; Gilula, NB (1986). "Cloning and characterization of human and rat liver cDNAs coding for a gap junction protein". The Journal of Cell Biology 103 (3): 767–76. doi:10.1083/jcb.103.3.767. PMID 2875078.
- ↑ "The ultrastructure of the nexus. A correlated thin-section and freeze-cleave study". J. Cell Biol. 47 (3): 666–88. December 1970. doi:10.1083/jcb.47.3.666. PMID 5531667.
- ↑ Chalcroft, J. P.; Bullivant, S (1970). "An interpretation of liver cell membrane and junction structure based on observation of freeze-fracture replicas of both sides of the fracture". The Journal of Cell Biology 47 (1): 49–60. doi:10.1083/jcb.47.1.49. PMID 4935338.
- ↑ Young; Cohn, ZA; Gilula, NB (1987). "Functional assembly of gap junction conductance in lipid bilayers: demonstration that the major 27 kd protein forms the junctional channel". Cell 48 (5): 733–43. doi:10.1016/0092-8674(87)90071-7. PMID 3815522.
- ↑ "The Mr 28,000 gap junction proteins from rat heart and liver are different but related". The Journal of Biological Chemistry 260 (11): 6514–6517. 1985. doi:10.1016/S0021-9258(18)88810-X. PMID 2987225.
- ↑ "Connexin43: a protein from rat heart homologous to a gap junction protein from liver". The Journal of Cell Biology 105 (6 Pt 1): 2621–2629. 1987. doi:10.1083/jcb.105.6.2621. PMID 2826492.
- ↑ "Identification of a 70,000-D protein in lens membrane junctional domains". The Journal of Cell Biology 101 (1): 28–35. 1985. doi:10.1083/jcb.101.1.28. PMID 3891760.
- ↑ "Three types of gap junctions interconnecting intestinal epithelial cells visualized by freeze-etching". Proc. Natl. Acad. Sci. U.S.A. 69 (5): 1318–21. May 1972. doi:10.1073/pnas.69.5.1318. PMID 4504340. Bibcode: 1972PNAS...69.1318S.
- ↑ Gruijters, WTM; Kistler, J; Bullivant, S; Goodenough, DA (1987). "Immunolocalization of MP70 in lens fiber 16-17-nm intercellular junctions". The Journal of Cell Biology 104 (3): 565–72. doi:10.1083/jcb.104.3.565. PMID 3818793.
- ↑ Robertson, JD (October 1963). "The occurrence of a subunit pattern in the unit membranes of club endings in mauthner cell synapses in goldfish brains". J. Cell Biol. 19 (1): 201–21. doi:10.1083/jcb.19.1.201. PMID 14069795.
- ↑ "Studies on an epithelial (gland) cell junction. I. Modifications of surface membrane permeability". J. Cell Biol. 22 (3): 565–86. September 1964. doi:10.1083/jcb.22.3.565. PMID 14206423.
- ↑ 150.0 150.1 Wong, Nai Sum, ed (2011). "The distribution of phosphatidylinositol 4,5-bisphosphate in acinar cells of rat pancreas revealed with the freeze-fracture replica labeling method". PLOS ONE 6 (8). doi:10.1371/journal.pone.0023567. PMID 21858170. Bibcode: 2011PLoSO...623567O.
- ↑ Strauss, RE; Gourdie, RG (December 2020). "Cx43 and the Actin Cytoskeleton: Novel Roles and Implications for Cell-Cell Junction-Based Barrier Function Regulation". Biomolecules 10 (12): 1656. doi:10.3390/biom10121656. PMID 33321985.
- ↑ Decker, RS; Friend, DS (July 1974). "Assembly of gap junctions during amphibian neurulation". J. Cell Biol. 62 (1): 32–47. doi:10.1083/jcb.62.1.32. PMID 4135001.
- ↑ Decker, RS (June 1976). "Hormonal regulation of gap junction differentiation". J. Cell Biol. 69 (3): 669–85. doi:10.1083/jcb.69.3.669. PMID 1083855.
- ↑ "Dynamic trafficking and delivery of connexons to the plasma membrane and accretion to gap junctions in living cells". Proc. Natl. Acad. Sci. U.S.A. 99 (16): 10446–51. August 2002. doi:10.1073/pnas.162055899. PMID 12149451. Bibcode: 2002PNAS...9910446L.
- ↑ Meyer, R; Malewicz, B; Baumann, WJ; Johnson, RG (June 1990). "Increased gap junction assembly between cultured cells upon cholesterol supplementation". J. Cell Sci. 96 (2): 231–8. doi:10.1242/jcs.96.2.231. PMID 1698798.
- ↑ Johnson, R. G.; Reynhout, J. K.; Tenbroek, E. M.; Quade, B. J.; Yasumura, T.; Davidson, K. G. V.; Sheridan, J. D.; Rash, J. E. (January 2012). "Gap junction assembly: roles for the formation plaque and regulation by the C-terminus of connexin43". Mol. Biol. Cell 23 (1): 71–86. doi:10.1091/mbc.E11-02-0141. PMID 22049024.
- ↑ Locke, Darren; Harris, Andrew L (2009). "Connexin channels and phospholipids: association and modulation". BMC Biol. 7 (1): 52. doi:10.1186/1741-7007-7-52. PMID 19686581.
- ↑ 158.0 158.1 "Connexin45-containing neuronal gap junctions in rodent retina also contain connexin36 in both apposing hemiplaques, forming bihomotypic gap junctions, with scaffolding contributed by zonula occludens-1". J. Neurosci. 28 (39): 9769–89. September 2008. doi:10.1523/JNEUROSCI.2137-08.2008. PMID 18815262.
Further reading
- Harris, Andrew; Locke, Darren, eds (2009). Connexins. New York: Springer. doi:10.1007/978-1-59745-489-6. ISBN 978-1-934115-46-6. https://www.springer.com/978-1-934115-46-6.
External links
- Gap+Junctions at the US National Library of Medicine Medical Subject Headings (MeSH)
 |


