Biology:Cation channel superfamily
| Ion channel (eukaryotic) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
 Potassium channel Kv1.2 (with beta2 auxiliary subunits), structure in a membrane-like environment. Calculated hydrocarbon boundaries of the lipid bilayer are indicated by red and blue dots. | |||||||||||
| Identifiers | |||||||||||
| Symbol | Ion_trans | ||||||||||
| Pfam | PF00520 | ||||||||||
| InterPro | IPR005821 | ||||||||||
| SCOP2 | 1bl8 / SCOPe / SUPFAM | ||||||||||
| TCDB | 1.A.1 | ||||||||||
| OPM superfamily | 8 | ||||||||||
| OPM protein | 2a79 | ||||||||||
| |||||||||||
| Ion channel (bacterial) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
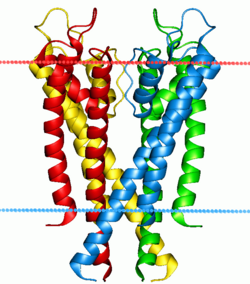 Potassium channel KcsA. Calculated hydrocarbon boundaries of the lipid bilayer are indicated by red and blue dots. | |||||||||||
| Identifiers | |||||||||||
| Symbol | Ion_trans_2 | ||||||||||
| Pfam | PF07885 | ||||||||||
| InterPro | IPR013099 | ||||||||||
| SCOP2 | 1bl8 / SCOPe / SUPFAM | ||||||||||
| OPM protein | 1r3j | ||||||||||
| |||||||||||
The transmembrane cation channel superfamily was defined in InterPro and Pfam as the family of tetrameric ion channels. These include the sodium, potassium,[1] calcium, ryanodine receptor, HCN, CNG, CatSper, and TRP channels. This large group of ion channels apparently includes families Template:TCDB, Template:TCDB, Template:TCDB, and Template:TCDB of the TCDB transporter classification.
They are described as minimally having two transmembrane helices flanking a loop which determines the ion selectivity of the channel pore. Many eukaryotic channels have four additional transmembrane helices (TM) (Pfam PF00520), related to or vestigial of voltage gating. The proteins with only two transmembrane helices (Pfam PF07885) are most commonly found in bacteria. This also includes the 2-TM inward-rectifier potassium channels (Pfam PF01007) found primarily in eukaryotes. There are commonly additional regulatory domains which serve to regulate ion conduction and channel gating. The pores may also be homotetramers or heterotetramers; where heterotetramers may be encoded as distinct genes or as multiple pore domains within a single polypeptide. The HVCN1 and Putative tyrosine-protein phosphatase proteins do not contain an expected ion conduction pore domain, but rather have homology only to the voltage sensor domain of voltage gated ion channels.
Human channels with 6 TM helices
Cation
Transient receptor potential
Canonical
Melastatin
Vanilloid
Mucolipin
- MCOLN1; MCOLN2; MCOLN3;
Ankyrin
TRPP
- PKD1L3;
Calcium
Voltage-dependent
Sperm
- CATSPER1; CATSPER2; CATSPER3; CATSPER4
Ryanodine receptor
Potassium
Voltage-gated potassium
Delayed rectifier
- Kvα1.x - Shaker-related: Kv1.1 (KCNA1), Kv1.2 (KCNA2), Kv1.3 (KCNA3), Kv1.5 (KCNA5), Kv1.6 (KCNA6), Kv1.7 (KCNA7), Kv1.8 (KCNA10)
- Kvα2.x - Shab-related: Kv2.1 (KCNB1), Kv2.2 (KCNB2)
- Kvα3.x - Shaw-related: Kv3.1 (KCNC1), Kv3.2 (KCNC2)
- Kvα7.x: Kv7.1 (KCNQ1) - KvLQT1, Kv7.2 (KCNQ2), Kv7.3 (KCNQ3), Kv7.4 (KCNQ4), Kv7.5 (KCNQ5)
- Kvα10.x: Kv10.1 (KCNH1)
A-type potassium
- Kvα1.x - Shaker-related: Kv1.4 (KCNA4)
- Kvα3.x - Shaw-related: Kv3.3 (KCNC3), Kv3.4 (KCNC4)
- Kvα4.x - Shal-related: Kv4.1 (KCND1), Kv4.2 (KCND2), Kv4.3 (KCND3)
Outward-rectifying
- Kvα10.x: Kv10.2 (KCNH5)
Inwardly-rectifying
Slowly activating
Modifier/silencer
- Kvα5.x: Kv5.1 (KCNF1)
- Kvα6.x: Kv6.1 (KCNG1), Kv6.2 (KCNG2), Kv6.3 (KCNG3), Kv6.4 (KCNG4)
- Kvα8.x: Kv8.1 (KCNV1), Kv8.2 (KCNV2)
- Kvα9.x: Kv9.1 (KCNS1), Kv9.2 (KCNS2), Kv9.3 (KCNS3)
Calcium-activated
BK
- KCa1.1 (BK, Slo1, Maxi-K, KCNMA1)
SK
- KCa2.x: KCa2.1 (KCNN1) - SK1, KCa2.2 (KCNN2) - SK2, KCa2.3 (KCNN3) - SK3
- KCa3.x: KCa3.1 (KCNN4) - SK4
- KCa4.x: KCa4.1 (KCNT1) - SLACK, KCa4.2 (KCNT2) - SLICK
IK
Other subfamilies
- KCa5.1 (Slo3, KCNU1)
Inward-rectifier potassium
Sodium
- NALCN
- SCN1A; SCN2A; SCN2A2; SCN3A; SCN4A; SCN5A; SCN7A; SCN8A; SCN9A; SCN10A; SCN11A
- SLC9A10; SLC9A11
Cyclic nucleotide-gated
Proton
Related proteins
- TPTE, part of the larger Voltage sensitive phosphatase family
Human channels with 2 TM helices in each subunit
Potassium
Tandem pore domain potassium channel
- KCNK1; KCNK2; KCNK3; KCNK4; KCNK5; KCNK6; KCNK7; KCNK9; KCNK10; KCNK12; KCNK13; KCNK15; KCNK16; KCNK17; KCNK18
Non-human channels
Two-pore
Pore-only potassium
Ligand-gated potassium
- GluR0[2]
Voltage-gated potassium
- KvAP[3]
Prokaryotic KCa
Voltage and cyclic nucleotide gated potassium
- MlotiK1[16]
Sodium
Non-selective
- NaK[22]
Prokaryotic inward-rectifier potassium
- KirBac[23]
Engineered
References
- ↑ "Potassium channel structures". Nature Reviews. Neuroscience 3 (2): 115–21. February 2002. doi:10.1038/nrn727. PMID 11836519.
- ↑ "Functional characterization of a potassium-selective prokaryotic glutamate receptor". Nature 402 (6763): 817–21. December 1999. doi:10.1038/45568. PMID 10617203. Bibcode: 1999Natur.402..817C.
- ↑ "X-ray structure of a voltage-dependent K+ channel". Nature 423 (6935): 33–41. May 2003. doi:10.1038/nature01580. PMID 12721618. Bibcode: 2003Natur.423...33J.
- ↑ "An Escherichia coli homologue of eukaryotic potassium channel proteins". Proceedings of the National Academy of Sciences of the United States of America 91 (9): 3510–4. April 1994. doi:10.1073/pnas.91.9.3510. PMID 8170937. Bibcode: 1994PNAS...91.3510M.
- ↑ "Structure of the RCK domain from the E. coli K+ channel and demonstration of its presence in the human BK channel". Neuron 29 (3): 593–601. March 2001. doi:10.1016/s0896-6273(01)00236-7. PMID 11301020.
- ↑ "Crystal structure and mechanism of a calcium-gated potassium channel". Nature 417 (6888): 515–22. May 2002. doi:10.1038/417515a. PMID 12037559. Bibcode: 2002Natur.417..515J.
- ↑ "Structural basis of allosteric interactions among Ca2+-binding sites in a K+ channel RCK domain". Nature Communications 4: 2621. 2013. doi:10.1038/ncomms3621. PMID 24126388. Bibcode: 2013NatCo...4.2621S.
- ↑ "Crystal structures of a ligand-free MthK gating ring: insights into the ligand gating mechanism of K+ channels". Cell 126 (6): 1161–73. September 2006. doi:10.1016/j.cell.2006.08.029. PMID 16990139.
- ↑ "Structure of the MthK RCK in complex with cadmium". Journal of Structural Biology 171 (2): 231–7. August 2010. doi:10.1016/j.jsb.2010.03.020. PMID 20371380.
- ↑ "Crystal structure of a Ba(2+)-bound gating ring reveals elementary steps in RCK domain activation". Structure 20 (12): 2038–47. December 2012. doi:10.1016/j.str.2012.09.014. PMID 23085076.
- ↑ "Crystal structure of a potassium ion transporter, TrkH". Nature 471 (7338): 336–40. March 2011. doi:10.1038/nature09731. PMID 21317882. Bibcode: 2011Natur.471..336C.
- ↑ "Gating of the TrkH ion channel by its associated RCK protein TrkA". Nature 496 (7445): 317–22. April 2013. doi:10.1038/nature12056. PMID 23598339. Bibcode: 2013Natur.496..317C.
- ↑ "The structure of the KtrAB potassium transporter". Nature 496 (7445): 323–8. April 2013. doi:10.1038/nature12055. PMID 23598340. Bibcode: 2013Natur.496..323V.
- ↑ "Distinct gating mechanisms revealed by the structures of a multi-ligand gated K(+) channel". eLife 1: e00184. December 2012. doi:10.7554/eLife.00184. PMID 23240087.
- ↑ "Crystal structure of a two-subunit TrkA octameric gating ring assembly". PLOS ONE 10 (3): e0122512. 2015. doi:10.1371/journal.pone.0122512. PMID 25826626. Bibcode: 2015PLoSO..1022512D.
- ↑ "Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel". Proceedings of the National Academy of Sciences of the United States of America 105 (5): 1511–5. February 2008. doi:10.1073/pnas.0711533105. PMID 18216238. Bibcode: 2008PNAS..105.1511C.
- ↑ "A prokaryotic voltage-gated sodium channel". Science 294 (5550): 2372–5. December 2001. doi:10.1126/science.1065635. PMID 11743207. Bibcode: 2001Sci...294.2372R.
- ↑ "The crystal structure of a voltage-gated sodium channel". Nature 475 (7356): 353–8. July 2011. doi:10.1038/nature10238. PMID 21743477.
- ↑ "Structure of a prokaryotic sodium channel pore reveals essential gating elements and an outer ion binding site common to eukaryotic channels". Journal of Molecular Biology 426 (2): 467–83. January 2014. doi:10.1016/j.jmb.2013.10.010. PMID 24120938. Bibcode: 2014BpJ...106..130A.
- ↑ "Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel". Nature 486 (7401): 130–4. May 2012. doi:10.1038/nature11054. PMID 22678295. Bibcode: 2012Natur.486..130Z.
- ↑ "Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing". Nature Communications 3: 1102. 2012. doi:10.1038/ncomms2077. PMID 23033078. Bibcode: 2012NatCo...3.1102M.
- ↑ "Atomic structure of a Na+- and K+-conducting channel". Nature 440 (7083): 570–4. March 2006. doi:10.1038/nature04508. PMID 16467789. Bibcode: 2006Natur.440..570S.
- ↑ "A family of putative Kir potassium channels in prokaryotes". BMC Evolutionary Biology 1: 14. 2001. doi:10.1186/1471-2148-1-14. PMID 11806753.
- ↑ "Tuning the ion selectivity of tetrameric cation channels by changing the number of ion binding sites". Proceedings of the National Academy of Sciences of the United States of America 108 (2): 598–602. January 2011. doi:10.1073/pnas.1013636108. PMID 21187421. Bibcode: 2011PNAS..108..598D.
- ↑ "Protein interactions central to stabilizing the K+ channel selectivity filter in a four-sited configuration for selective K+ permeation". Proceedings of the National Academy of Sciences of the United States of America 108 (40): 16634–9. October 2011. doi:10.1073/pnas.1111688108. PMID 21933962. Bibcode: 2011PNAS..10816634S.
External links
- "Voltage-gated Ion Channels". IUPHAR Database of Receptors and Ion Channels. International Union of Basic and Clinical Pharmacology. http://www.iuphar-db.org/IC/ReceptorFamiliesForward.
 |

