Biology:Cytochrome c oxidase
| Cytochrome c oxidase | |||||||||
|---|---|---|---|---|---|---|---|---|---|
 | |||||||||
| Identifiers | |||||||||
| EC number | 1.9.3.1 | ||||||||
| CAS number | 9001-16-5 | ||||||||
| Databases | |||||||||
| IntEnz | IntEnz view | ||||||||
| BRENDA | BRENDA entry | ||||||||
| ExPASy | NiceZyme view | ||||||||
| KEGG | KEGG entry | ||||||||
| MetaCyc | metabolic pathway | ||||||||
| PRIAM | profile | ||||||||
| PDB structures | RCSB PDB PDBe PDBsum | ||||||||
| Gene Ontology | AmiGO / QuickGO | ||||||||
| |||||||||
| Cytochrome c oxidase | |
|---|---|
 | |
| Identifiers | |
| Symbol | Cytochrome c oxidase |
| OPM superfamily | 4 |
| OPM protein | 2dyr |
| Membranome | 257 |
The enzyme cytochrome c oxidase or Complex IV, (was EC 1.9.3.1, now reclassified as a translocase EC 7.1.1.9) is a large transmembrane protein complex found in bacteria, archaea, and the mitochondria of eukaryotes.[1]
It is the last enzyme in the respiratory electron transport chain of cells located in the membrane. It receives an electron from each of four cytochrome c molecules and transfers them to one oxygen molecule and four protons, producing two molecules of water. In addition to binding the four protons from the inner aqueous phase, it transports another four protons across the membrane, increasing the transmembrane difference of proton electrochemical potential, which the ATP synthase then uses to synthesize ATP.
Structure
The complex
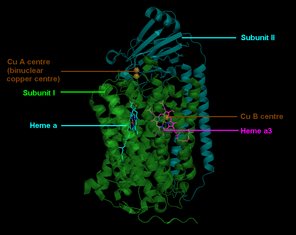
The complex is a large integral membrane protein composed of several metal prosthetic sites and 14[2] protein subunits in mammals. In mammals, eleven subunits are nuclear in origin, and three are synthesized in the mitochondria. The complex contains two hemes, a cytochrome a and cytochrome a3, and two copper centers, the CuA and CuB centers.[3] In fact, the cytochrome a3 and CuB form a binuclear center that is the site of oxygen reduction. Cytochrome c, which is reduced by the preceding component of the respiratory chain (cytochrome bc1 complex, Complex III), docks near the CuA binuclear center and passes an electron to it, being oxidized back to cytochrome c containing Fe3+. The reduced CuA binuclear center now passes an electron on to cytochrome a, which in turn passes an electron on to the cytochrome a3>-CuB binuclear center. The two metal ions in this binuclear center are 4.5 Å apart and coordinate a hydroxide ion in the fully oxidized state.
Crystallographic studies of cytochrome c oxidase show an unusual post-translational modification, linking C6 of Tyr(244) and the ε-N of His(240) (bovine enzyme numbering). It plays a vital role in enabling the cytochrome a3- CuB binuclear center to accept four electrons in reducing molecular oxygen and four protons to water. The mechanism of reduction was formerly thought to involve a peroxide intermediate, which was believed to lead to superoxide production. However, the currently accepted mechanism involves a rapid four-electron reduction involving immediate oxygen–oxygen bond cleavage, avoiding any intermediate likely to form superoxide.[4]:865–866
The conserved subunits
| No. | Subunit name | Human protein | Protein description from UniProt | Pfam family with Human protein |
|---|---|---|---|---|
| 1 | Cox1 | COX1 HUMAN | Cytochrome c oxidase subunit 1 | Pfam PF00115 |
| 2 | Cox2 | COX2 HUMAN | Cytochrome c oxidase subunit 2 | Pfam PF02790, Pfam PF00116 |
| 3 | Cox3 | COX3 HUMAN | Cytochrome c oxidase subunit 3 | Pfam PF00510 |
| 4 | Cox4i1 | COX41 HUMAN | Cytochrome c oxidase subunit 4 isoform 1, mitochondrial | Pfam PF02936 |
| 5 | Cox4a2 | COX42 HUMAN | Cytochrome c oxidase subunit 4 isoform 2, mitochondrial | Pfam PF02936 |
| 6 | Cox5a | COX5A HUMAN | Cytochrome c oxidase subunit 5A, mitochondrial | Pfam PF02284 |
| 7 | Cox5b | COX5B HUMAN | Cytochrome c oxidase subunit 5B, mitochondrial | Pfam PF01215 |
| 8 | Cox6a1 | CX6A1 HUMAN | Cytochrome c oxidase subunit 6A1, mitochondrial | Pfam PF02046 |
| 9 | Cox6a2 | CX6A2 HUMAN | Cytochrome c oxidase subunit 6A2, mitochondrial | Pfam PF02046 |
| 10 | Cox6b1 | CX6B1 HUMAN | Cytochrome c oxidase subunit 6B1 | Pfam PF02297 |
| 11 | Cox6b2 | CX6B2 HUMAN | Cytochrome c oxidase subunit 6B2 | Pfam PF02297 |
| 12 | Cox6c | COX6C HUMAN | Cytochrome c oxidase subunit 6C | Pfam PF02937 |
| 13 | Cox7a1 | CX7A1 HUMAN | Cytochrome c oxidase subunit 7A1, mitochondrial | Pfam PF02238 |
| 14 | Cox7a2 | CX7A2 HUMAN | Cytochrome c oxidase subunit 7A2, mitochondrial | Pfam PF02238 |
| 15 | Cox7a3 | COX7S HUMAN | Putative cytochrome c oxidase subunit 7A3, mitochondrial | Pfam PF02238 |
| 16 | Cox7b | COX7B HUMAN | Cytochrome c oxidase subunit 7B, mitochondrial | Pfam PF05392 |
| 17 | Cox7c | COX7C HUMAN | Cytochrome c oxidase subunit 7C, mitochondrial | Pfam PF02935 |
| 18 | Cox7r | COX7R HUMAN | Cytochrome c oxidase subunit 7A-related protein, mitochondrial | Pfam PF02238 |
| 19 | Cox8a | COX8A HUMAN | Cytochrome c oxidase subunit 8A, mitochondrial P | Pfam PF02285 |
| 20 | Cox8c | COX8C HUMAN | Cytochrome c oxidase subunit 8C, mitochondrial | Pfam PF02285 |
| 1 | Coa1 | COA1 HUMAN | Cytochrome c oxidase assembly factor 1 homolog | Pfam PF08695 |
| 2 | Coa3 | COA3 HUMAN | Cytochrome c oxidase assembly factor 3 homolog, mitochondrial | Pfam PF09813 |
| 3 | Coa4 | COA4 HUMAN | Cytochrome c oxidase assembly factor 4 homolog, mitochondrial | Pfam PF06747 |
| 4 | Coa5 | COA5 HUMAN | Cytochrome c oxidase assembly factor 5 | Pfam PF10203 |
| 5 | Coa6 | COA6 HUMAN | Cytochrome c oxidase assembly factor 6 homolog | Pfam PF02297 |
| 6 | Coa7 | COA7 HUMAN | Cytochrome c oxidase assembly factor 7, | Pfam PF08238 |
| 7 | Cox11 | COX11 HUMAN | Cytochrome c oxidase assembly protein COX11 mitochondrial | Pfam PF04442 |
| 8 | Cox14 | COX14 HUMAN | Cytochrome c oxidase assembly protein | Pfam PF14880 |
| 9 | Cox15 | COX15 HUMAN | Cytochrome c oxidase assembly protein COX15 homolog | Pfam PF02628 |
| 10 | Cox16 | COX16 HUMAN | Cytochrome c oxidase assembly protein COX16 homolog mitochondrial | Pfam PF14138 |
| 11 | Cox17 | COX17 HUMAN | Cytochrome c oxidase copper chaperone | Pfam PF05051 |
| 12 | Cox18[7] | COX18 HUMAN | Mitochondrial inner membrane protein (Cytochrome c oxidase assembly protein 18) | Pfam PF02096 |
| 13 | Cox19 | COX19 HUMAN | Cytochrome c oxidase assembly protein | Pfam PF06747 |
| 14 | Cox20 | COX20 HUMAN | Cytochrome c oxidase protein 20 homolog | Pfam PF12597 |
Assembly
COX assembly in yeast are a complex process that is not entirely understood due to the rapid and irreversible aggregation of hydrophobic subunits that form the holoenzyme complex, as well as aggregation of mutant subunits with exposed hydrophobic patches.[8] COX subunits are encoded in both the nuclear and mitochondrial genomes. The three subunits that form the COX catalytic core are encoded in the mitochondrial genome. Over 30 different nuclear-encoded chaperone proteins are required for COX assembly.[9]
Cofactors, including hemes, are inserted into subunits I & II. The two heme molecules reside in subunit I, helping with transport to subunit II where two copper molecules aid with the continued transfer of electrons.[10] Subunits I and IV initiate assembly. Different subunits may associate to form sub-complex intermediates that later bind to other subunits to form the COX complex.[8] In post-assembly modifications, COX will form a homodimer. This is required for activity. Dimers are connected by a cardiolipin molecule,[8][11][12] which has been found to play a key role in stabilization of the holoenzyme complex. The dissociation of subunits VIIa and III in conjunction with the removal of cardiolipin results in total loss of enzyme activity.[12] Subunits encoded in the nuclear genome are known to play a role in enzyme dimerization and stability. Mutations to these subunits eliminate COX function.[8]
Assembly is known to occur in at least three distinct rate-determining steps. The products of these steps have been found, though specific subunit compositions have not been determined.[8]
Synthesis and assembly of COX subunits I, II, and III are facilitated by translational activators, which interact with the 5’ untranslated regions of mitochondrial mRNA transcripts. Translational activators are encoded in the nucleus. They can operate through either direct or indirect interaction with other components of translation machinery, but exact molecular mechanisms are unclear due to difficulties associated with synthesizing translation machinery in-vitro.[13][14] Though the interactions between subunits I, II, and III encoded within the mitochondrial genome make a lesser contribution to enzyme stability than interactions between bigenomic subunits, these subunits are more conserved, indicating potential unexplored roles for enzyme activity.[15]
Biochemistry
This section is missing information about names of the six traditional intermediate states (APFOER); 2021 Cyro-EM result proposing an RPFOE mechanism with reversed assignment of red-ox phases (doi:10.1038/s41467-021-27174-y |
The overall reaction is
- 4 Fe2+ – cytochrome c + 4 H+ + O2 → 4 Fe3+ – cytochrome c + 2 H2O ΔfGo' = - 218 kJ/mol
Two electrons are passed from two cytochrome c's, through the CuA and cytochrome a sites to the cytochrome a3–CuB binuclear center, reducing the metals to the Fe2+ form and Cu+. The hydroxide ligand is protonated and lost as water, creating a void between the metals that is filled by O2. The oxygen is rapidly reduced, with two electrons coming from the Fe2+-cytochrome a3, which is converted to the ferryl oxo form (Fe4+=O). The oxygen atom close to CuB picks up one electron from Cu+, and a second electron and a proton from the hydroxyl of Tyr(244), which becomes a tyrosyl radical. The second oxygen is converted to a hydroxide ion by picking up two electrons and a proton. A third electron from another cytochrome c is passed through the first two electron carriers to the cytochrome a3–CuB binuclear center, and this electron and two protons convert the tyrosyl radical back to Tyr, and the hydroxide bound to CuB2+ to a water molecule. The fourth electron from another cytochrome c flows through CuA and cytochrome a to the cytochrome a3–CuB binuclear center, reducing the Fe4+=O to Fe3+, with the oxygen atom picking up a proton simultaneously, regenerating this oxygen as a hydroxide ion coordinated in the middle of the cytochrome a3–CuB center as it was at the start of this cycle. Overall, four reduced cytochrome c's are oxidized while O2 and four protons are reduced to two water molecules.[4]:841–5
Inhibition
COX exists in three conformational states: fully oxidized (pulsed), partially reduced, and fully reduced. Each inhibitor has a high affinity to a different state. In the pulsed state, both the heme a3 and the CuB nuclear centers are oxidized; this is the conformation of the enzyme that has the highest activity. A two-electron reduction initiates a conformational change that allows oxygen to bind at the active site to the partially-reduced enzyme. Four electrons bind to COX to fully reduce the enzyme. Its fully reduced state, which consists of a reduced Fe2+ at the cytochrome a3 heme group and a reduced CuB+ binuclear center, is considered the inactive or resting state of the enzyme.[16]
Cyanide, azide, and carbon monoxide[17] all bind to cytochrome c oxidase, inhibiting the protein from functioning and leading to the chemical asphyxiation of cells. Higher concentrations of molecular oxygen are needed to compensate for increasing inhibitor concentrations, leading to an overall decrease in metabolic activity in the cell in the presence of an inhibitor. Other ligands, such as nitric oxide and hydrogen sulfide, can also inhibit COX by binding to regulatory sites on the enzyme, reducing the rate of cellular respiration.[18]
Cyanide is a non-competitive inhibitor for COX,[19][20] binding with high affinity to the partially-reduced state of the enzyme and hindering further reduction of the enzyme. In the pulsed state, cyanide binds slowly, but with high affinity. The ligand is posited to electrostatically stabilize both metals at once by positioning itself between them. A high nitric oxide concentration, such as one added exogenously to the enzyme, reverses cyanide inhibition of COX.[21]
Nitric oxide can reversibly[22] bind to either metal ion in the binuclear center to be oxidized to nitrite. NO and CN− will compete with oxygen to bind at the site, reducing the rate of cellular respiration. Endogenous NO, however, which is produced at lower levels, augments CN− inhibition. Higher levels of NO, which correlate with the existence of more enzyme in the reduced state, lead to a greater inhibition of cyanide.[16] At these basal concentrations, NO inhibition of Complex IV is known to have beneficial effects, such as increasing oxygen levels in blood vessel tissues. The inability of the enzyme to reduce oxygen to water results in a buildup of oxygen, which can diffuse deeper into surrounding tissues.[22] NO inhibition of Complex IV has a larger effect at lower oxygen concentrations, increasing its utility as a vasodilator in tissues of need.[22]
Hydrogen sulfide will bind COX in a noncompetitive fashion at a regulatory site on the enzyme, similar to carbon monoxide. Sulfide has the highest affinity to either the pulsed or partially reduced states of the enzyme, and is capable of partially reducing the enzyme at the heme a3 center. It is unclear whether endogenous H2S levels are sufficient to inhibit the enzyme. There is no interaction between hydrogen sulfide and the fully reduced conformation of COX.[18]
Methanol in methylated spirits is converted into formic acid, which also inhibits the same oxidase system. High levels of ATP can allosterically inhibit cytochrome c oxidase, binding from within the mitochondrial matrix.[23]
Extramitochondrial and subcellular localizations
Cytochrome c oxidase has 3 subunits which are encoded by mitochondrial DNA (cytochrome c oxidase subunit I, subunit II, and subunit III). Of these 3 subunits encoded by mitochondrial DNA, two have been identified in extramitochondrial locations. In pancreatic acinar tissue, these subunits were found in zymogen granules. Additionally, in the anterior pituitary, relatively high amounts of these subunits were found in growth hormone secretory granules.[24] The extramitochondrial function of these cytochrome c oxidase subunits has not yet been characterized. Besides cytochrome c oxidase subunits, extramitochondrial localization has also been observed for large numbers of other mitochondrial proteins.[25][26] This raises the possibility about existence of yet unidentified specific mechanisms for protein translocation from mitochondria to other cellular destinations.[24][26][27]
Genetic defects and disorders
Defects involving genetic mutations altering cytochrome c oxidase (COX) functionality or structure can result in severe, often fatal metabolic disorders. Such disorders usually manifest in early childhood and affect predominantly tissues with high energy demands (brain, heart, muscle). Among the many classified mitochondrial diseases, those involving dysfunctional COX assembly are thought to be the most severe.[28]
The vast majority of COX disorders are linked to mutations in nuclear-encoded proteins referred to as assembly factors, or assembly proteins. These assembly factors contribute to COX structure and functionality, and are involved in several essential processes, including transcription and translation of mitochondrion-encoded subunits, processing of preproteins and membrane insertion, and cofactor biosynthesis and incorporation.[29]
Currently, mutations have been identified in seven COX assembly factors: SURF1, SCO1, SCO2, COX10, COX15, COX20, COA5 and LRPPRC. Mutations in these proteins can result in altered functionality of sub-complex assembly, copper transport, or translational regulation. Each gene mutation is associated with the etiology of a specific disease, with some having implications in multiple disorders. Disorders involving dysfunctional COX assembly via gene mutations include Leigh syndrome, cardiomyopathy, leukodystrophy, anemia, and sensorineural deafness.
Histochemistry
The increased reliance of neurons on oxidative phosphorylation for energy[30] facilitates the use of COX histochemistry in mapping regional brain metabolism in animals, since it establishes a direct and positive correlation between enzyme activity and neuronal activity.[31] This can be seen in the correlation between COX enzyme amount and activity, which indicates the regulation of COX at the level of gene expression. COX distribution is inconsistent across different regions of the animal brain, but its pattern of its distribution is consistent across animals. This pattern has been observed in the monkey, mouse, and calf brain. One isozyme of COX has been consistently detected in histochemical analysis of the brain.[32] Such brain mapping has been accomplished in spontaneous mutant mice with cerebellar disease such as reeler[33] and a transgenic model of Alzheimer's disease.[34] This technique has also been used to map learning activity in the animal brain.[35]
Additional images
See also
- Cytochrome c oxidase subunit I
- Cytochrome c oxidase subunit II
- Cytochrome c oxidase subunit III
- Heme a
References
- ↑ "Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen" (in en). The EMBO Journal 13 (11): 2516–2525. June 1994. doi:10.1002/j.1460-2075.1994.tb06541.x. PMID 8013452.
- ↑ "NDUFA4 is a subunit of complex IV of the mammalian electron transport chain". Cell Metabolism 16 (3): 378–86. September 2012. doi:10.1016/j.cmet.2012.07.015. PMID 22902835.
- ↑ "Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 A". Science 269 (5227): 1069–74. August 1995. doi:10.1126/science.7652554. PMID 7652554. Bibcode: 1995Sci...269.1069T.
- ↑ 4.0 4.1 Voet, Donald; Voet, Judith G. (2011). Biochemistry (4th ed.). Hoboken, NJ: John Wiley & Sons. ISBN 978-0-470-57095-1.
- ↑ "Electron transfer by domain movement in cytochrome bc1". Nature 392 (6677): 677–84. April 1998. doi:10.1038/33612. PMID 9565029. Bibcode: 1998Natur.392..677Z.
- ↑ "A combined quantum chemical and crystallographic study on the oxidized binuclear center of cytochrome c oxidase". Biochimica et Biophysica Acta (BBA) - Bioenergetics 1807 (7): 769–78. July 2011. doi:10.1016/j.bbabio.2010.12.016. PMID 21211513.
- ↑ "The COX18 gene, involved in mitochondrial biogenesis, is functionally conserved and tightly regulated in humans and fission yeast". FEMS Yeast Research 6 (6): 869–82. September 2006. doi:10.1111/j.1567-1364.2006.00083.x. PMID 16911509.
- ↑ 8.0 8.1 8.2 8.3 8.4 "Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process". American Journal of Physiology. Cell Physiology 291 (6): C1129-47. December 2006. doi:10.1152/ajpcell.00233.2006. PMID 16760263.
- ↑ Dickinson, Elizabeth K.; Adams, Denise L.; Schon, Eric A.; Glerum, D. Moira (September 2000). "A Human SCO2 Mutation Helps Define the Role of Sco1p in the Cytochrome Oxidase Assembly Pathway" (in en). Journal of Biological Chemistry 275 (35): 26780–26785. doi:10.1016/S0021-9258(19)61443-2.
- ↑ Crofts, Antony (1996). "Cytochrome oxidase: Complex IV". University of Illinois at Urbana-Champaign. http://www.life.illinois.edu/crofts/bioph354/cyt_ox.html.
- ↑ "Biogenesis of cytochrome c oxidase". Mitochondrion 5 (6): 363–88. December 2005. doi:10.1016/j.mito.2005.08.002. PMID 16199211.
- ↑ 12.0 12.1 "Destabilization of the Quaternary Structure of Bovine Heart Cytochrome c Oxidase upon Removal of Tightly Bound Cardiolipin". Biochemistry 54 (36): 5569–77. September 2015. doi:10.1021/acs.biochem.5b00540. PMID 26284624.
- ↑ "Control of protein synthesis in yeast mitochondria: the concept of translational activators". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1833 (2): 286–94. February 2013. doi:10.1016/j.bbamcr.2012.03.007. PMID 22450032.
- ↑ "Biogenesis and assembly of eukaryotic cytochrome c oxidase catalytic core". Biochimica et Biophysica Acta (BBA) - Bioenergetics 1817 (6): 883–97. June 2012. doi:10.1016/j.bbabio.2011.09.005. PMID 21958598.
- ↑ "Protein-protein interfaces from cytochrome c oxidase I evolve faster than nonbinding surfaces, yet negative selection is the driving force". Genome Biology and Evolution 6 (11): 3064–76. October 2014. doi:10.1093/gbe/evu240. PMID 25359921.
- ↑ 16.0 16.1 "Interaction of cyanide and nitric oxide with cytochrome c oxidase: implications for acute cyanide toxicity". Toxicological Sciences 101 (1): 101–11. January 2008. doi:10.1093/toxsci/kfm254. PMID 17906319.
- ↑ "Carbon monoxide specifically inhibits cytochrome c oxidase of human mitochondrial respiratory chain". Pharmacology & Toxicology 93 (3): 142–6. September 2003. doi:10.1034/j.1600-0773.2003.930306.x. PMID 12969439.
- ↑ 18.0 18.1 "Sulfide inhibition of and metabolism by cytochrome c oxidase". Biochemical Society Transactions 41 (5): 1312–6. October 2013. doi:10.1042/BST20130070. PMID 24059525.
- ↑ Roberts, Michael; Reiss, Michael Jonathan; Monger, Grace (2000) (in en). Advanced Biology. Nelson Thornes. ISBN 9780174387329. https://books.google.com/books?id=HHaDGynAz1EC&q=cyanide+cytochrome+competitive&pg=PA130. Retrieved 2020-10-25.
- ↑ (in en) Biology: A Functional Approach. Nelson Thornes. 1986. ISBN 9780174480198. https://books.google.com/books?id=ASADBUVAiDUC&q=cyanide+cytochrome+competitive&pg=PA92. Retrieved 2020-10-25.
- ↑ "Cyanide inhibition of cytochrome c oxidase. A rapid-freeze e.p.r. investigation". The Biochemical Journal 224 (3): 829–37. December 1984. doi:10.1042/bj2240829. PMID 6098268.
- ↑ 22.0 22.1 22.2 "The ligand binding battle at cytochrome c oxidase: how NO regulates oxygen gradients in tissue". Circulation Research 104 (10): 1136–8. May 2009. doi:10.1161/CIRCRESAHA.109.198911. PMID 19461104.
- ↑ "Cell respiration s controlled by ATP, an allosteric inhibitor of cytochrome-c oxidase.". Eur J Biochem 249 (1): 350–354. October 1997. doi:10.1111/j.1432-1033.1997.t01-1-00350.x. PMID 9363790.
- ↑ 24.0 24.1 "Localization of mitochondrial DNA encoded cytochrome c oxidase subunits I and II in rat pancreatic zymogen granules and pituitary growth hormone granules". Histochemistry and Cell Biology 124 (5): 409–21. November 2005. doi:10.1007/s00418-005-0056-2. PMID 16133117.
- ↑ "Unusual Cellular Disposition of the Mitochondrial Molecular Chaperones Hsp60, Hsp70 and Hsp10". The Biology of Extracellular Molecular Chaperones. Novartis Foundation Symposia. 291. 2008. 59–68; discussion 69–73, 137–40. doi:10.1002/9780470754030.ch5. ISBN 9780470754030.
- ↑ 26.0 26.1 "Mitochondrial proteins at unexpected cellular locations: export of proteins from mitochondria from an evolutionary perspective". International Review of Cytology 194: 133–96. 1999. doi:10.1016/S0074-7696(08)62396-7. ISBN 9780123645982. PMID 10494626.
- ↑ "Mitochondrial-matrix proteins at unexpected locations: are they exported?". Trends in Biochemical Sciences 24 (5): 174–7. May 1999. doi:10.1016/s0968-0004(99)01390-0. PMID 10322429.
- ↑ "Genetic defects of cytochrome c oxidase assembly". Physiological Research 53 (Suppl 1): S213-23. 2004. doi:10.33549/physiolres.930000.53.S213. PMID 15119951. http://www.biomed.cas.cz/physiolres/pdf/53%20Suppl%201/53_S213.pdf. Retrieved 2010-11-17.
- ↑ "Defects in cytochrome oxidase assembly in humans: lessons from yeast". Biochemistry and Cell Biology 84 (6): 859–69. December 2006. doi:10.1139/o06-201. PMID 17215873.
- ↑ "Neuron-specific specificity protein 4 bigenomically regulates the transcription of all mitochondria- and nucleus-encoded cytochrome c oxidase subunit genes in neurons". Journal of Neurochemistry 127 (4): 496–508. November 2013. doi:10.1111/jnc.12433. PMID 24032355.
- ↑ "Cytochrome oxidase: an endogenous metabolic marker for neuronal activity". Trends in Neurosciences 12 (3): 94–101. March 1989. doi:10.1016/0166-2236(89)90165-3. PMID 2469224.
- ↑ "Brain cytochrome oxidase: purification, antibody production, and immunohistochemical/histochemical correlations in the CNS". The Journal of Neuroscience 9 (11): 3884–98. November 1989. doi:10.1523/jneurosci.09-11-03884.1989. PMID 2555458.
- ↑ "Regional brain variations of cytochrome oxidase activity in Relnrl-orl mutant mice". Journal of Neuroscience Research 83 (5): 821–31. April 2006. doi:10.1002/jnr.20772. PMID 16511878.
- ↑ "Regional brain cytochrome oxidase activity in beta-amyloid precursor protein transgenic mice with the Swedish mutation". Neuroscience 118 (4): 1151–63. 2003. doi:10.1016/S0306-4522(03)00037-X. PMID 12732258.
- ↑ "Spatial learning of the water maze: progression of brain circuits mapped with cytochrome oxidase histochemistry". Neurobiology of Learning and Memory 93 (3): 362–71. March 2010. doi:10.1016/j.nlm.2009.12.002. PMID 19969098.
External links
- The Cytochrome Oxidase home page at Rice University
- Interactive Molecular model of cytochrome c oxidase (Requires MDL Chime)
- UMich Orientation of Proteins in Membranes families/superfamily-4
- Cytochrome-c+Oxidase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |




