Biology:Mitochondrial membrane transport protein
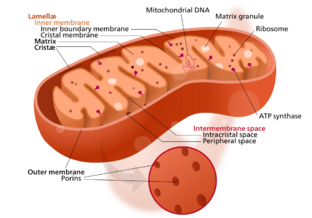
Mitochondrial membrane transport proteins, also known as mitochondrial carrier proteins, are proteins which exist in the membranes of mitochondria. They serve to transport[2] molecules and other factors, such as ions, into or out of the organelles. Mitochondria contain both an inner and outer membrane, separated by the inter-membrane space, or inner boundary membrane. The outer membrane is porous, whereas the inner membrane restricts the movement of all molecules. The two membranes also vary in membrane potential and pH.[3] These factors play a role in the function of mitochondrial membrane transport proteins. There are 53 discovered human mitochondrial membrane transporters,[4] with many others that are known to still need discovered.
Mitochondrial outer membrane
The outer mitochondrial membrane forms the border of mitochondria towards the cellular environment. The outer membrane mitochondrial proteins carry out functions for mitochondrial biogenesis and integration between mitochondria and the cellular system. The outer membrane consists of two types of integral proteins, including proteins with transmembrane β-barrel and proteins with one or more α-helical membrane anchors.[5][6]
β-Barrel Outer Membrane Proteins
TOM complex
The TOM complex, part of the TOM/TIM supercomplex, is essential for the translocase of almost all mitochondrial proteins which consists of at least 7 different subunits. Tom20 and Tom70 are the primary receptors while Tom40, Tom22, Tom7, Tom6, and Tom5 subunits form the stable TOM Complex.[7][8][9] The receptor proteins Tom70 and Tom20 recognize incoming precursor proteins, in which Tom70 is responsible for docking of precursors of hydrophobic proteins accompanied by cytosolic chaperones and Tom 20 recognizes precursor proteins of the presequence pathways.[10][11][12][13][14][15][16] Tom40 is the protein-conducting channel of the complex with beta-barrel structure,[17][18] which forms a cation-selective channel. Tom40 has a large pore diameter of 22Å that can allow the accommodation of partially folded protein structure[19] The inner wall of Tom40 has a charged region that allows interaction with hydrophilic precursor proteins while the hydrophobic precursor of ADP/ATP carrier can be crosslinked with the hydrophobic region of Tom40. Three small proteins Tom5, Tom6, Tom7 interact closely with Tom40 to assemble and stabilize the complex. The TOM complex also consists of a dimer of Tom40 or small Tom proteins that are held together by two Tom22 subunits.[20][21] Protein sorting into the mitochondrial compartments always starts at the TOM complex. The TOM complex forms two exit sites for precursor proteins—Tom40, Tom7, and the intermembrane space domain of Tom22—promote the transfer of presequence-containing precursors to the TIM23 complex.[20]
SAM complex
The SAM Complex is essential for sorting and assembling beta-barrel proteins from the intermembrane space side into the outer membrane.[22][23][24] The SAM complex consists of three subunits: The β-barrel protein Sam50 and two peripheral subunits Sam35 and Sam37.[22][25][26] Sam50 belongs to the conserved Omp85 protein family which can be characterized by a 16-stranded β-barrel and by a different number of polypeptide transport-associated (POTRA) domains.[23][24] Sam50 exposes a single POTRA domain towards the intermembrane space.[26][27] Sam35 caps the Sam50 β-barrel, stabilizing the core of the protein translocase.[26][28][29] Sam50 and Sam35 are responsible for the binding of precursors of β-barrel proteins, which contain conserved β-signal that is formed by the last β-strand.[30][31] The β-barrel of Sam50 is the functional domain that inserts and folds substrate proteins into the outer membrane.
Sam35 binds to Sam50 and closely interacts with Sam37, in which Sam37 does not bind to Sam50. Sam37 and Sam35 have a conformation similar to glutathione-S-transferase, except they do not possess residues required for enzymatic activity. Sam37 accommodates the release of the folded β-barrel proteins from the SAM complex.[31]
Voltage-dependent anion ion channel or VDAC
VDAC (voltage-dependent anion ion channel) is important for the exchange of small hydrophilic ions and metabolites with the cytosol, which is driven by the gradient concentration across the outer membrane. VDAC is the most abundant protein in the outer membrane.[32][33] Like Tom40, VDAC has a β-barrel structure with antiparallel β-strands that can facilitate the passage of β-barrel membrane proteins. VDAC has a pore size of 2-4 nm for small hydrophilic molecules. VDAC plays a crucial role in facilitating energy metabolism by transporting ADP and ATP in and out of the outer membrane. VDAC also accommodates the passage of NADH and many anionic metabolites. VDAC operation is voltage-dependent in which it closes at high voltage and can partially open towards slightly reduced anion selectivity.[34][35]
α-Helical outer membrane proteins
The Mitochondrial import complex (MIM)
The import pathways of α-helical membrane anchors or signal-anchored proteins are carried out mainly by outer membrane proteins.[6] Precursors of the polytopic or multi-spanning proteins can be recognized by Tom70, but cannot be passed through the Tom40 channel.[12][36][37] Tom70 transfers the precursor proteins to the MIM Complex. The MIM complex constitutes the major inserts for alpha-helical proteins into the target membrane.[12][13][37] The MIM Complex consists of several copies of Mim1 and one or two copies of Mim2. Both subunits are necessary for stabilizing partner proteins and for outer membrane protein biogenesis[38]
Mitochondrial inner membrane
The inner mitochondrial membrane is a structure that surrounds the mitochondrial matrix, characterized by many folds and compartments that form crista and is the site of oxidative phosphorylation and ATP synthesis.[3][39] The high concentration of cardiolipin, a type of lipid and about 20% of the inner membrane composition, makes it impermeable to most molecules. Specialized transporters arranged in specific configurations are required to regulate the diffusion of molecules across the membrane. The inner membrane's structure causes a membrane potential of approximately 180 mV.[39]
Respiratory chain supercomplex

The respiratory chain supercomplex is located in the cristae of the inner membrane. It is composed of multiple complexes that work together to drive oxidative phosphorylation and ATP synthesis. The complexes cannot function without the other parts of the respiratory supercomplex being present.[39] The supercomplex is the site of the mitochondrial electron transport chain.[40]
NADH/ubiquinone oxidoreductase
NADH/ubiquinone oxidoreductase, also known as complex I, is the first and largest protein in the mitochondrial respiratory chain. It consists of a membrane arm, embedded inside the inner mitochondrial membrane, and a matrix arm, extending out of the membrane. There are 78 transmembrane helices and three proton pumps. The junction of the two arms is the site of conduction of NADH to ubiquinol.[39] Complex I is a scaffold needed for complex III and IV, and it will not function without these other complexes being present.[40]
Cytochrome c reductase, succinate dehydrogenase, and cytochrome c oxidase
Cytochrome c reductase, also known as complex III, is the second protein in the respiratory chain. It pumps electrons from complex I, through succinate dehydrogenase (complex II) to cytochrome c (complex IV). Complex III and IV are proton pumps, pumping H+ protons out of the mitochondrial matrix, and work in conjunction with complex I to create the proton gradient found at the inner membrane. Cytochrome c is and electron carrier protein that travels between complex III and IV, and triggers apoptosis if it leaves the cristae. Complex IV passes electrons to oxygen, the final acceptor in the mitochondrial electron transport chain.[40][3]
Inner membrane translocases
TIM complex
The TIM complex is a protein translocase located on the inner membrane. It is part of the TOM/TIM supercomplex, which spans the intermembrane space.[3] The TIM complex is responsible for sorting proteins into the mitochondrial matrix or into the membrane. TIM22 and TIM23 are the main subunits. TIM 22 is responsible for allowing other mitochondrial transporters to insert themselves into the inner membrane, whereas TIM23 reads proteins with an N-terminus precursor for import into the membrane or matrix.[41]
ADP, ATP translocase
ADP, ATP translocase is responsible for regulating the movement of ADP and ATP in and out of the inner membrane. ATP is sorted into the cytosol, while ADP is sorted into the mitochondrial matrix to undergo oxidative phosphorylation. Due to the constant demand of ATP production, ADP, ATP translocases are in higher abundance than other transporters.[42][43] ADP, ATP translocase is a small protein, ~30-33 kDa, composed of 6 transmembrane α-helices, that form 3 repeat domains for an overall funnel-like structure in the membrane. Towards the center of the funnel structure it has a 7 amino acid loop 12. It is structurally unique when compared to other proteins that interact with ATP in that it lacks adenosine monophosphate and requires at least two phosphate groups to allow for passage of the molecule. It's composed of 297 amino acid residues, with 18 of them being charged molecules. The ADP, ATP translocase is opened in the presence of Ca2+.[43][39]
Phosphate transport proteins
Phosphate transport proteins are similar in structure and are both part of the same family of mitochondrial carriers. It consists of 6 transmembrane α-helices, but lacks the 7 amino acid loop 12 found in ADP, ATP translocase. Phosphate transport proteins are responsible for transport of phosphate across the inner membrane so it can be used in the phosphorylation of ADP.[39]
Mutations of mitochondrial membrane transporters
Mutations of DNA coding for mitochondrial membrane transport proteins are linked to a wide range of diseases and disorders, such as cardiomyopathy, encephalopathy, muscular dystrophy, epilepsy, neuropathy, and fingernail dysplasia.[44] Most mutations of mitochondrial membrane transporters are autosomal recessive. Mutations to transporters within the inner mitochondrial membrane mostly affect high-energy tissues due to the disruption of oxidative phosphorylation.[4][44] For example, decreased mitochondrial function has been linked to heart failure and hypertrophy. This mitochondrial response translates into a shift towards glycolysis and lactate production that can cause tumor formation and proliferation of the tissues.[40]
Examples
Examples of mitochondrial transport proteins include the following:
- The mitochondrial permeability transition pore, which opens in response to increased mitochondrial calcium (Ca2+) load and oxidative stress[45]
- The mitochondrial calcium uniporter which transports calcium from the cytosol of the cell into the mitochondrial matrix[45][46]
- The mitochondrial sodium/calcium exchanger, which carries Ca2+ ions out of the matrix in exchange for Na+ ions. These transport proteins serve to maintain the proper electrical and chemical gradients in mitochondria by keeping ions and other factors in the right balance between the inside and outside of mitochondria.
See also
References
- ↑ "File:Mitochondrion structure.svg" (in en), Wikipedia, https://en.wikipedia.org/wiki/File:Mitochondrion_structure.svg, retrieved 2021-05-03
- ↑ Mitochondrial+Membrane+Transport+Proteins at the US National Library of Medicine Medical Subject Headings (MeSH)
- ↑ 3.0 3.1 3.2 3.3 3.4 "Structure and function of mitochondrial membrane protein complexes". BMC Biology 13 (1): 89. October 2015. doi:10.1186/s12915-015-0201-x. PMID 26515107.
- ↑ 4.0 4.1 "The SLC25 Carrier Family: Important Transport Proteins in Mitochondrial Physiology and Pathology". Physiology 35 (5): 302–327. September 2020. doi:10.1152/physiol.00009.2020. PMID 32783608.
- ↑ "Definition of a High-Confidence Mitochondrial Proteome at Quantitative Scale". Cell Reports 19 (13): 2836–2852. June 2017. doi:10.1016/j.celrep.2017.06.014. PMID 28658629.
- ↑ 6.0 6.1 "Proteomic analysis of the yeast mitochondrial outer membrane reveals accumulation of a subclass of preproteins". Molecular Biology of the Cell 17 (3): 1436–50. March 2006. doi:10.1091/mbc.e05-08-0740. PMID 16407407.
- ↑ "Preprotein translocase of the outer mitochondrial membrane: molecular dissection and assembly of the general import pore complex". Molecular and Cellular Biology 18 (11): 6515–24. November 1998. doi:10.1128/mcb.18.11.6515. PMID 9774667.
- ↑ "The preprotein translocation channel of the outer membrane of mitochondria". Cell 93 (6): 1009–19. June 1998. doi:10.1016/s0092-8674(00)81206-4. PMID 9635430.
- ↑ "The TOM core complex: the general protein import pore of the outer membrane of mitochondria". The Journal of Cell Biology 147 (5): 959–68. November 1999. doi:10.1083/jcb.147.5.959. PMID 10579717.
- ↑ "Molecular chaperones Hsp90 and Hsp70 deliver preproteins to the mitochondrial import receptor Tom70". Cell 112 (1): 41–50. January 2003. doi:10.1016/s0092-8674(02)01250-3. PMID 12526792.
- ↑ "Roles of Tom70 in import of presequence-containing mitochondrial proteins" (in English). The Journal of Biological Chemistry 284 (46): 31635–46. November 2009. doi:10.1074/jbc.M109.041756. PMID 19767391.
- ↑ 12.0 12.1 12.2 "The mitochondrial import protein Mim1 promotes biogenesis of multispanning outer membrane proteins". The Journal of Cell Biology 194 (3): 387–95. August 2011. doi:10.1083/jcb.201102044. PMID 21825073.
- ↑ 13.0 13.1 "Multispan mitochondrial outer membrane protein Ugo1 follows a unique Mim1-dependent import pathway". The Journal of Cell Biology 194 (3): 397–405. August 2011. doi:10.1083/jcb.201102041. PMID 21825074.
- ↑ "Recruitment of Cytosolic J-Proteins by TOM Receptors Promotes Mitochondrial Protein Biogenesis". Cell Reports 25 (8): 2036–2043.e5. November 2018. doi:10.1016/j.celrep.2018.10.083. PMID 30463002.
- ↑ "Tom70 enhances mitochondrial preprotein import efficiency by binding to internal targeting sequences". The Journal of Cell Biology 217 (4): 1369–1382. April 2018. doi:10.1083/jcb.201708044. PMID 29382700.
- ↑ "Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import". The Journal of Biological Chemistry 283 (7): 3799–807. February 2008. doi:10.1074/jbc.m708339200. PMID 18063580.
- ↑ "Detection of likely transmembrane beta strand regions in sequences of mitochondrial pore proteins using the Gibbs sampler". Journal of Bioenergetics and Biomembranes 28 (2): 163–9. April 1996. doi:10.1007/bf02110647. PMID 9132415.
- ↑ "Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins [see comment]". Nature 395 (6701): 516–21. October 1998. doi:10.1038/26780. PMID 9774109.
- ↑ The three modules of ADP/ATP carrier cooperate in receptor recruitment and translocation into mitochondria. Oxford University Press. 2001-03-01. OCLC 678227688. http://worldcat.org/oclc/678227688.
- ↑ 20.0 20.1 "Structure of the mitochondrial import gate reveals distinct preprotein paths". Nature 575 (7782): 395–401. November 2019. doi:10.1038/s41586-019-1680-7. PMID 31600774.
- ↑ "Cryo-EM structure of the mitochondrial protein-import channel TOM complex at near-atomic resolution". Nature Structural & Molecular Biology 26 (12): 1158–1166. December 2019. doi:10.1038/s41594-019-0339-2. PMID 31740857. PMC 8439582. https://escholarship.org/uc/item/0mc942j6.
- ↑ 22.0 22.1 "Machinery for protein sorting and assembly in the mitochondrial outer membrane". Nature 424 (6948): 565–71. July 2003. doi:10.1038/nature01753. PMID 12891361.
- ↑ 23.0 23.1 "The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria". The Journal of Cell Biology 164 (1): 19–24. January 2004. doi:10.1083/jcb.200310092. PMID 14699090.
- ↑ 24.0 24.1 "Membrane protein insertion through a mitochondrial β-barrel gate". Science 359 (6373): eaah6834. January 2018. doi:10.1126/science.aah6834. PMID 29348211.
- ↑ "Characterization of the insertase for β-barrel proteins of the outer mitochondrial membrane". The Journal of Cell Biology 199 (4): 599–611. November 2012. doi:10.1083/jcb.201207161. PMID 23128244.
- ↑ 26.0 26.1 26.2 "Structural insight into mitochondrial β-barrel outer membrane protein biogenesis". Nature Communications 11 (1): 3290. July 2020. doi:10.1038/s41467-020-17144-1. PMID 32620929.
- ↑ "The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial beta-barrel proteins". The Journal of Cell Biology 176 (1): 77–88. January 2007. doi:10.1083/jcb.200602050. PMID 17190789.
- ↑ "Tob38, a novel essential component in the biogenesis of beta-barrel proteins of mitochondria". EMBO Reports 5 (7): 704–9. July 2004. doi:10.1038/sj.embor.7400183. PMID 15205677.
- ↑ "Sam35 of the mitochondrial protein sorting and assembly machinery is a peripheral outer membrane protein essential for cell viability". The Journal of Biological Chemistry 279 (21): 22781–5. May 2004. doi:10.1074/jbc.c400120200. PMID 15067005.
- ↑ "Dissecting membrane insertion of mitochondrial beta-barrel proteins". Cell 132 (6): 1011–24. March 2008. doi:10.1016/j.cell.2008.01.028. PMID 18358813.
- ↑ 31.0 31.1 "The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis". Molecular Biology of the Cell 19 (1): 126–36. January 2008. doi:10.1091/mbc.e07-08-0796. PMID 17978093.
- ↑ "Voltage-dependent anion channels: the wizard of the mitochondrial outer membrane". Biological Chemistry 395 (12): 1435–42. December 2014. doi:10.1515/hsz-2014-0203. PMID 25153596.
- ↑ "Revisiting trends on mitochondrial mega-channels for the import of proteins and nucleic acids". Journal of Bioenergetics and Biomembranes 49 (1): 75–99. February 2017. doi:10.1007/s10863-016-9662-z. PMID 27146409.
- ↑ "Mitochondrial outer membrane channels". Chemical Reviews 112 (12): 6373–87. December 2012. doi:10.1021/cr3002033. PMID 22979903.
- ↑ "Solution structure of the integral human membrane protein VDAC-1 in detergent micelles". Science 321 (5893): 1206–10. August 2008. doi:10.1126/science.1161302. PMID 18755977.
- ↑ "A novel insertion pathway of mitochondrial outer membrane proteins with multiple transmembrane segments". The Journal of Cell Biology 179 (7): 1355–63. December 2007. doi:10.1083/jcb.200702143. PMID 18158327.
- ↑ 37.0 37.1 "Mitochondrial protein translocation-associated degradation". Nature 569 (7758): 679–683. May 2019. doi:10.1038/s41586-019-1227-y. PMID 31118508.
- ↑ "A crucial role for Mim2 in the biogenesis of mitochondrial outer membrane proteins". Journal of Cell Science 125 (Pt 14): 3464–73. July 2012. doi:10.1242/jcs.103804. PMID 22467864.
- ↑ 39.0 39.1 39.2 39.3 39.4 39.5 "Transport proteins (carriers) of mitochondria". IUBMB Life 61 (1): 40–6. January 2009. doi:10.1002/iub.139. PMID 18816452.
- ↑ 40.0 40.1 40.2 40.3 "The function of the respiratory supercomplexes: the plasticity model". Biochimica et Biophysica Acta (BBA) - Bioenergetics 1837 (4): 444–50. April 2014. doi:10.1016/j.bbabio.2013.12.009. PMID 24368156.
- ↑ "Protein translocation into mitochondria: the role of TIM complexes". Trends in Cell Biology 10 (1): 25–31. January 2000. doi:10.1016/S0962-8924(99)01684-0. PMID 10603473.
- ↑ "Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside". Nature 426 (6962): 39–44. November 2003. doi:10.1038/nature02056. PMID 14603310.
- ↑ 43.0 43.1 "The ADP and ATP transport in mitochondria and its carrier". Biochimica et Biophysica Acta (BBA) - Biomembranes 1778 (10): 1978–2021. October 2008. doi:10.1016/j.bbamem.2008.04.011. PMID 18510943.
- ↑ 44.0 44.1 "Diseases Caused by Mutations in Mitochondrial Carrier Genes SLC25: A Review". Biomolecules 10 (4): 655. April 2020. doi:10.3390/biom10040655. PMID 32340404.
- ↑ 45.0 45.1 "The mitochondrial permeability transition pore and its role in cell death". The Biochemical Journal 341 (Pt 2): 233–49. July 1999. doi:10.1042/bj3410233. PMID 10393078.
- ↑ "Mitochondria and calcium signaling". Cell Calcium 38 (3–4): 311–7. 2005. doi:10.1016/j.ceca.2005.06.011. PMID 16087232.
 |

