Biology:mir-7 microRNA precursor
| mir-7 microRNA precursor | |
|---|---|
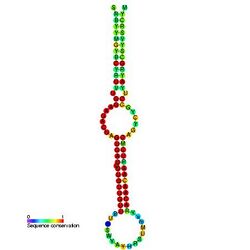 Predicted secondary structure and sequence conservation of mir-7 | |
| Identifiers | |
| Symbol | mir-7 |
| Rfam | RF00053 |
| miRBase | MI0000263 |
| miRBase family | MIPF0000022 |
| Other data | |
| RNA type | Gene; miRNA |
| Domain(s) | Eukaryota |
| GO | 0035195 0035068 |
| SO | 0001244 |
| PDB structures | PDBe |
This family represents the microRNA (miRNA) precursor mir-7. This miRNA has been predicted or experimentally confirmed in a wide range of species.[1] miRNAs are transcribed as ~70 nucleotide precursors (modelled here) and subsequently processed by the Dicer enzyme to give a ~22 nucleotide product. In this case the mature sequence comes from the 5' arm of the precursor. The extents of the hairpin precursors are not generally known and are estimated based on hairpin prediction. The involvement of Dicer in miRNA processing suggests a relationship with the phenomenon of RNA interference.
Mature miRNA-7 is derived from three microRNA precursors in the human genome, miR-7-1, miR-7-2 and miR-7-3. miRNAs are numbered based on the sequence of the mature RNA.
miR-7 is directly regulated by the transcription factor HoxD10.[2]
miRNAs are thought to have regulatory roles through complementarity to mRNA. miR-7 is essential for the maintenance of regulatory stability under conditions of environmental flux.[3] It plays an important role in controlling mRNA expression. The miR-7 gene is found in most sequenced Urbilateria species, and the sequence of its mature miRNA product is perfectly conserved from annelids to humans, indicating a strong functional conservation.[3]
Targets of miR-7
Bioinformatic predictions suggest that the human EGFR mRNA 3'-untranslated region contains three microRNA-7 (miR-7) target sites, which are not conserved across mammals.[4] In Drosophila photoreceptor cells, miR-7 controls epidermal growth factor receptor (EGFR) signaling and promotes photoreceptor differentiation.[5] Among other targets of miR-7 are insulin-like growth factor 1 receptor (IGF1R) and PIK3CD,[6] E(spl) gene family [7] and Pak1 (cancer cells).[2] c-Fos is also a target of miR-7b in mice.[8] Pax6 translation in the lateral wall of the subventricular zone of developed mice is post-transcriptionally regulated by miRNA-7a mediated gene silencing, which is necessary to control the rate of dopaminergic neuron production in the olfactory bulb.[9]
Clinical relevance
Multiple roles and targets of miR-7 as well as its expression pattern were linked to regulatory mechanisms and pathogenesis in glioblastoma,[10] breast cancer[11] and other types of cancers,[4][6][12] as well as in schizophrenia[13] and visual abnormalities.[14] Inhibition of the motility, invasiveness, anchorage-independent growth, and tumorigenic potential of highly invasive breast cancer cells through the introduction of miR-7 suggests a strong therapeutic potential of miR-7.[2][15]
References
- ↑ "miRNA gene family: mir-7 (92 sequences)". MiRBase. http://microrna.sanger.ac.uk/cgi-bin/sequences/mirna_summary.pl?fam=MIPF0000022.
- ↑ 2.0 2.1 2.2 "MicroRNA-7, a homeobox D10 target, inhibits p21-activated kinase 1 and regulates its functions". Cancer Research 68 (20): 8195–200. October 2008. doi:10.1158/0008-5472.CAN-08-2103. PMID 18922890.
- ↑ 3.0 3.1 "A microRNA imparts robustness against environmental fluctuation during development". Cell 137 (2): 273–82. April 2009. doi:10.1016/j.cell.2009.01.058. PMID 19379693.
- ↑ 4.0 4.1 "Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7". The Journal of Biological Chemistry 284 (9): 5731–41. February 2009. doi:10.1074/jbc.M804280200. PMID 19073608.
- ↑ "A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye". Cell 123 (7): 1267–77. December 2005. doi:10.1016/j.cell.2005.10.040. PMID 16377567.
- ↑ 6.0 6.1 "MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells". The Biochemical Journal 432 (1): 199–205. November 2010. doi:10.1042/BJ20100859. PMID 20819078.
- ↑ "Identification of Drosophila MicroRNA targets". PLOS Biology 1 (3): E60. December 2003. doi:10.1371/journal.pbio.0000060. PMID 14691535.

- ↑ "miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation". Proceedings of the National Academy of Sciences of the United States of America 103 (42): 15669–74. October 2006. doi:10.1073/pnas.0605781103. PMID 17028171.
- ↑ "miR-7a regulation of Pax6 controls spatial origin of forebrain dopaminergic neurons". Nature Neuroscience 15 (8): 1120–1126. 2012. doi:10.1038/nn.3142. PMID 22729175.
- ↑ "microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma". Cancer Research 68 (10): 3566–72. May 2008. doi:10.1158/0008-5472.CAN-07-6639. PMID 18483236.
- ↑ "Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer". Proceedings of the National Academy of Sciences of the United States of America 105 (35): 13021–6. September 2008. doi:10.1073/pnas.0803304105. PMID 18755890. Bibcode: 2008PNAS..10513021F.
- ↑ "MiRNA expression in urothelial carcinomas: important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31". International Journal of Cancer 124 (9): 2236–42. May 2009. doi:10.1002/ijc.24183. PMID 19127597.
- ↑ "microRNA expression in the prefrontal cortex of individuals with schizophrenia and schizoaffective disorder". Genome Biology 8 (2): R27. 2007. doi:10.1186/gb-2007-8-2-r27. PMID 17326821.
- ↑ "Prediction and verification of miRNA expression in human and rat retinas". Investigative Ophthalmology & Visual Science 48 (9): 3962–7. September 2007. doi:10.1167/iovs.06-1221. PMID 17724173.
- ↑ "MicroRNAs as therapeutic targets". The New England Journal of Medicine 354 (11): 1194–5. March 2006. doi:10.1056/NEJMcibr060065. PMID 16540623.
Further reading
- "Identification of novel genes coding for small expressed RNAs". Science 294 (5543): 853–8. October 2001. doi:10.1126/science.1064921. PMID 11679670. Bibcode: 2001Sci...294..853L.
- "microRNAs: tiny regulators with great potential". Cell 107 (7): 823–6. December 2001. doi:10.1016/S0092-8674(01)00616-X. PMID 11779458.
- "miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro". Biochemical and Biophysical Research Communications 394 (4): 921–7. April 2010. doi:10.1016/j.bbrc.2010.03.076. PMID 20230785.
- "Post-transcriptional regulation of alpha-synuclein expression by mir-7 and mir-153". The Journal of Biological Chemistry 285 (17): 12726–34. April 2010. doi:10.1074/jbc.M109.086827. PMID 20106983.
- "Drosophila maelstrom ensures proper germline stem cell lineage differentiation by repressing microRNA-7". Developmental Cell 17 (3): 417–24. September 2009. doi:10.1016/j.devcel.2009.07.017. PMID 19758565.
- "Repression of alpha-synuclein expression and toxicity by microRNA-7". Proceedings of the National Academy of Sciences of the United States of America 106 (31): 13052–7. August 2009. doi:10.1073/pnas.0906277106. PMID 19628698. Bibcode: 2009PNAS..10613052J.
- "MicroRNA miR-7 is preferentially expressed in endocrine cells of the developing and adult human pancreas". Gene Expression Patterns 9 (4): 193–9. April 2009. doi:10.1016/j.gep.2008.12.003. PMID 19135553.
External links
- Page for mir-7 microRNA precursor at Rfam
- Purow, Benjamin (December 2008). "MIR7-1 (microRNA 7-1)". Atlas of Genetics and Cytogenetics in Oncology and Haematology. http://atlasgeneticsoncology.org/Genes/MIRN71ID44356ch9q21.html.
 |

