Biology:Galactokinase
| Galactokinase 1 | |
|---|---|
 | |
| Identifiers | |
| Symbol | GALK1 |
| Alt. symbols | GALK |
| NCBI gene | 2584 |
| HGNC | 4118 |
| OMIM | 604313 |
| RefSeq | NM_000154 |
| UniProt | P51570 |
| Other data | |
| EC number | 2.7.1.6 |
| Locus | Chr. 17 q23-q25 |
| Galactokinase 2 | |
|---|---|
| Identifiers | |
| Symbol | GALK2 |
| NCBI gene | 2585 |
| HGNC | 4119 |
| OMIM | 137028 |
| RefSeq | NM_002044 |
| UniProt | Q01415 |
| Other data | |
| EC number | 2.7.1.6 |
| Locus | Chr. 15 [1] |
Galactokinase is an enzyme (phosphotransferase) that facilitates the phosphorylation of α-D-galactose to galactose 1-phosphate at the expense of one molecule of ATP.[1] Galactokinase catalyzes the second step of the Leloir pathway, a metabolic pathway found in most organisms for the catabolism of α-D-galactose to glucose 1-phosphate.[2] First isolated from mammalian liver, galactokinase has been studied extensively in yeast,[3][4] archaea,[5] plants,[6][7] and humans.[8][9]
Structure
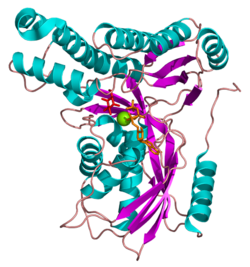
Galactokinase is composed of two domains separated by a large cleft. The two regions are known as the N- and C-terminal domains, and the adenine ring of ATP binds in a hydrophobic pocket located at their interface. The N-terminal domain is marked by five strands of mixed beta-sheet and five alpha-helices, and the C-terminal domain is characterized by two layers of anti-parallel beta-sheets and six alpha-helices.[8] Galactokinase does not belong to the sugar kinase family, but rather to a class of ATP-dependent enzymes known as the GHMP superfamily.[10] GHMP is an abbreviation referring to its original members: galactokinase, homoserine kinase, mevalonate kinase, and phosphomevalonate kinase. Members of the GHMP superfamily have great three-dimensional similarity despite only ten to 20% sequence identity. These enzymes contain three well-conserved motifs (I, II, and III), the second of which is involved in nucleotide binding and has the sequence Pro-X-X-X-Gly-Leu-X-Ser-Ser-Ala.[11]
Sugar specificity
Galactokinases across different species display a great diversity of substrate specificities. E. coli galactokinase can also phosphorylate 2-deoxy-D-galactose, 2-amino-deoxy-D-galactose, 3-deoxy-D-galactose and D-fucose. The enzyme cannot tolerate any C-4 modifications, but changes at the C-2 position of D-galactose do not interfere with enzyme function.[12] Both human and rat galactokinases are also able to successfully phosphorylate 2-deoxy-D-galactose.[13][14] Galactokinase from S. cerevisiae, on the other hand, is highly specific for D-galactose and cannot phosphorylate glucose, mannose, arabinose, fucose, lactose, galactitol, or 2-deoxy-D-galactose.[3][4] Moreover, the kinetic properties of galactokinase also differ across species.[8] The sugar specificity of galactokinases from different sources has been dramatically expanded through directed evolution[15] and structure-based protein engineering.[16][17] The corresponding broadly permissive sugar anomeric kinases serve as a cornerstone for in vitro and in vivo glycorandomization.[18][19][20]
Mechanism
Recently, the roles of active site residues in human galactokinase have become understood. Asp-186 abstracts a proton from C1-OH of α-D-galactose, and the resulting alkoxide nucleophile attacks the γ-phosphorus of ATP. A phosphate group is transferred to the sugar, and Asp-186 may be deprotonated by water. Nearby Arg-37 stabilizes Asp-186 in its anionic form and has also been proven to be essential to galactokinase function in point mutation experiments.[9] Both the aspartic acid and arginine active site residues are highly conserved among galactokinases.[8]


Biological function
The Leloir pathway catalyzes the conversion of galactose to glucose. Galactose is found in dairy products, as well as in fruits and vegetables, and can be produced endogenously in the breakdown of glycoproteins and glycolipids. Three enzymes are required in the Leloir pathway: galactokinase, galactose-1-phosphate uridylyltransferase, and UDP-galactose 4-epimerase. Galactokinase catalyzes the first committed step of galactose catabolism, forming galactose 1-phosphate.[2][21]
Disease relevance
Galactosemia, a rare metabolic disorder characterized by decreased ability to metabolize galactose, can be caused by a mutation in any of the three enzymes in the Leloir pathway.[2] Galactokinase deficiency, also known as galactosemia type II, is a recessive metabolic disorder caused by a mutation in human galactokinase. About 20 mutations have been identified that cause galactosemia type II, the main symptom of which is early onset cataracts. In lens cells of the human eye, aldose reductase converts galactose to galactitol. As galactose is not being catabolized to glucose due to a galactokinase mutation, galactitol accumulates. This galactitol gradient across the lens cell membrane triggers the osmotic uptake of water, and the swelling and eventual apoptosis of lens cells ensues.[22]
References
- ↑ "galactokinase". Medical Dictionary. http://www.theodora.com/medical_dictionary/gamma_galactosis.html#galactokinase.
- ↑ 2.0 2.1 2.2 "The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose". FASEB Journal 10 (4): 461–70. March 1996. doi:10.1096/fasebj.10.4.8647345. PMID 8647345.
- ↑ 3.0 3.1 "Purification of galactokinase mRNA from Saccharomyces cerevisiae by indirect immunoprecipitation". The Journal of Biological Chemistry 254 (9): 3531–6. May 1979. doi:10.1016/S0021-9258(18)50793-6. PMID 107173.
- ↑ 4.0 4.1 "Contribution of amino acid side chains to sugar binding specificity in a galactokinase, Gal1p, and a transcriptional inducer, Gal3p". The Journal of Biological Chemistry 281 (25): 17150–5. June 2006. doi:10.1074/jbc.M602086200. PMID 16603548.
- ↑ "Substrate specificity and mechanism from the structure of Pyrococcus furiosus galactokinase". Journal of Molecular Biology 337 (2): 387–98. March 2004. doi:10.1016/j.jmb.2004.01.043. PMID 15003454.
- ↑ "[Purification and mechanism of action of a plant galactokinase]". Biochimie 58 (5): 499–504. 1976. doi:10.1016/s0300-9084(76)80218-0. PMID 182286.
- ↑ "Galactokinase of Vicia faba seeds". European Journal of Biochemistry 136 (1): 155–9. October 1983. doi:10.1111/j.1432-1033.1983.tb07720.x. PMID 6617655.
- ↑ 8.0 8.1 8.2 8.3 "Galactokinase: structure, function and role in type II galactosemia". Cellular and Molecular Life Sciences 61 (19–20): 2471–84. October 2004. doi:10.1007/s00018-004-4160-6. PMID 15526155.
- ↑ 9.0 9.1 9.2 "The role of the active site residues in human galactokinase: implications for the mechanisms of GHMP kinases". Bioorganic Chemistry 39 (3): 120–6. June 2011. doi:10.1016/j.bioorg.2011.03.001. PMID 21474160.
- ↑ "Molecular and biochemical characterization of human galactokinase and its small molecule inhibitors". Chemico-Biological Interactions 188 (3): 376–85. December 2010. doi:10.1016/j.cbi.2010.07.025. PMID 20696150.
- ↑ 11.0 11.1 "Molecular structure of galactokinase". The Journal of Biological Chemistry 278 (35): 33305–11. August 2003. doi:10.1074/jbc.M304789200. PMID 12796487.
- ↑ "Studies on the substrate specificity of Escherichia coli galactokinase". Organic Letters 5 (13): 2223–6. June 2003. doi:10.1021/ol034642d. PMID 12816414.
- ↑ "Sugar recognition by human galactokinase". BMC Biochemistry 4: 16. November 2003. doi:10.1186/1471-2091-4-16. PMID 14596685.
- ↑ "Some properties of galactokinase in developing rat liver". The Biochemical Journal 108 (2): 169–75. June 1968. doi:10.1042/bj1080169. PMID 5665881.
- ↑ "Creation of the first anomeric D/L-sugar kinase by means of directed evolution". Proceedings of the National Academy of Sciences of the United States of America 100 (23): 13184–9. November 2003. doi:10.1073/pnas.2235011100. PMID 14612558.
- ↑ "Structure-based engineering of E. coli galactokinase as a first step toward in vivo glycorandomization". Chemistry & Biology 12 (6): 657–64. June 2005. doi:10.1016/j.chembiol.2005.04.009. PMID 15975511.
- ↑ "The impact of enzyme engineering upon natural product glycodiversification". Current Opinion in Chemical Biology 12 (5): 556–64. October 2008. doi:10.1016/j.cbpa.2008.07.013. PMID 18678278.
- ↑ "Neoglycorandomization and chemoenzymatic glycorandomization: two complementary tools for natural product diversification". Journal of Natural Products 68 (11): 1696–711. November 2005. doi:10.1021/np0502084. PMID 16309329.
- ↑ "Recombinant E. coli prototype strains for in vivo glycorandomization". ACS Chemical Biology 6 (1): 95–100. January 2011. doi:10.1021/cb100267k. PMID 20886903.
- ↑ "Enzymatic methods for glyco(diversification/randomization) of drugs and small molecules". Natural Product Reports 28 (11): 1811–53. October 2011. doi:10.1039/c1np00045d. PMID 21901218.
- ↑ "Structure and function of enzymes of the Leloir pathway for galactose metabolism". The Journal of Biological Chemistry 278 (45): 43885–8. November 2003. doi:10.1074/jbc.R300025200. PMID 12923184.
- ↑ "Functional analysis of disease-causing mutations in human galactokinase". European Journal of Biochemistry 270 (8): 1767–74. April 2003. doi:10.1046/j.1432-1033.2003.03538.x. PMID 12694189.
External links
- Galactokinase at the US National Library of Medicine Medical Subject Headings (MeSH)
 |

