Biology:miR-155
| pre-mir-155 | |
|---|---|
 pre-mir-155 secondary structure and sequence conservation.ard | |
| Identifiers | |
| Symbol | miR-155 |
| Rfam | RF00731 |
| miRBase family | MIPF0000157 |
| Other data | |
| RNA type | microRNA |
| Domain(s) | Eukaryota; |
| PDB structures | PDBe |
 Generic protein structure example |
MiR-155 is a microRNA that in humans is encoded by the MIR155 host gene or MIR155HG.[1] MiR-155 plays a role in various physiological and pathological processes.[2][3][4][5][6][7] Exogenous molecular control in vivo of miR-155 expression may inhibit malignant growth,[8][9] viral infections,[10] and enhance the progression of cardiovascular diseases.[11]
Discovery
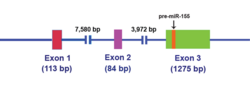
The MIR155HG was initially identified as a gene that was transcriptionally activated by promoter insertion at a common retroviral integration site in B-cell lymphomas and was formerly called BIC (B-cell Integration Cluster). The MIR155HG is transcribed by RNA polymerase II and the resulting ~1,500 nucleotide RNA is capped and polyadenylated. The 23 nucleotide single-stranded miR-155, which is harbored in exon 3, is subsequently processed from the parent RNA molecule.[12]
Biogenesis
The MIR155HG RNA transcript does not contain a long open reading frame (ORF), however, it does include an imperfectly base-paired stem loop that is conserved across species.[13] This non-coding RNA (ncRNA) is now defined as a primary-miRNA (pri-miRNA).[13] Once miR-155 pri-miRNA is transcribed, this transcript is cleaved by the nuclear microprocessor complex, of which the core components are the RNase III type endonuclease Drosha and the DiGeorge critical region 8 (DGCR8) protein,[14][15] to produce a 65 nucleotide stem-loop precursor miRNA (pre-mir-155) (see Figure 2).

Following export from the nucleus by exportin-5, pre-mir-155 molecules are cleaved near the terminal loop by Dicer resulting in RNA duplexes of ~22nucleotides.[14][15] Following Dicer cleavage, an Argonaute (Ago) protein binds to the short RNA duplexes, forming the core of a multi-subunit complex called the RNA-induced silencing complex (RISC).[16] In a manner similar to siRNA duplexes, one of the two strands, the "passenger miRNA" (miR-155*), is released and degraded while the other strand, designated the "guide strand" or "mature miRNA" (miR-155), is retained within the RISC.[16]
Recent data suggest that both arms of the pre-miRNA hairpin can give rise to mature miRNAs.[17][18] Due to the increasing number of examples where two functional mature miRNAs are processed from opposite arms of the same pre-miRNA, pre-mir-155 products are now denoted with the suffix -5p (from the 5′ arm) (e.g. miR-155-5p) and -3p (from the 3′ arm) (e.g. miR-155-3p) following their name (see Figure 3).[19]

Once miR-155-5p/-3p is assembled into the RISC, these molecules subsequently recognize their target messenger RNA (mRNA) by base pairing interactions between nucleotides 2 and 8 of miR-155-5p/-3p (the seed region) and complementary nucleotides predominantly in the 3′-untranslated region (3′-UTR) of mRNAs (see Figure 4 and 5 below).[20] Finally, with the miR-155-5p/-3p acting as an adaptor for the RISC, complex-bound mRNAs are subjected to translational repression (i.e. inhibition of translation initiation) and/or degradation following deadenylation.[16]
Evolutionary conservation
Early phylogenetic analyses demonstrated that the sequence of pre-mir-155 and miR-155-5p was conserved between human, mouse, and chicken.[13] Recent annotated sequencing data found that 22 different organisms including, mammals, amphibians, birds, reptiles, sea squirts, and sea lampreys, express a conserved miR-155-5p.[1] Currently much less sequence data is available regarding miR-155-3p, therefore, it is not clear how conserved this miRNA is across species.[2]
Tissue distribution
Northern blot analysis found that miR-155 pri-miRNA was abundantly expressed in the human spleen and thymus and detectable in the liver, lung, and kidney.[13] Subsequently, polymerase chain reaction (PCR) experiments demonstrated that miR-155-5p was detectable in all human tissues investigated.[21] Sequence analysis of small RNA clone libraries comparing miRNA expression to all other organ systems examined established that miR-155-5p was one of five miRNAs (i.e. miR-142, miR-144, miR-150, miR-155, and miR-223) that was specific for hematopoietic cells including B-cells, T-cells, monocytes and granulocytes.[22] Together these results suggest that miR-155-5p is expressed in a number of tissues and cell types and, therefore, may play a critical role in a wide variety of biological processes, including hematopoiesis[2][3][4]
Although very few studies have investigated the expression levels of miR-155-3p, Landgraf et al.[22] established that expression levels of this miRNA was very low in hematopoietic cells. Additionally, PCR analyses found that while miR-155-3p was detectable in a number of human tissues the expression levels of this miRNA were 20–200 fold less when compared to miR-155-5p levels.[23] Even though the function of miR-155-3p has been largely ignored, several studies now suggest that, in some cases (astrocytes and plasmacytoid dendritic cells), both miR-155-5p and -3p can be functionally matured from pre-mir-155.[24][25]
Targets
Bioinformatic analysis using TargetScan 6.2 (release date June, 2012) [3] revealed at least 4,174 putative human miR-155-5p mRNA targets exist, with a total of 918 conserved sites (i.e. between mouse and human) and 4,249 poorly conserved sites (i.e. human only).[20][26] Although the TargetScan 6.2 algorithm cannot be utilized to determine the miR-155-3p putative targets, one would speculate that this miRNA may also potentially regulate the expression of thousands of mRNA targets.
A comprehensive list of miR-155-5p/mRNA targets that were experimentally authenticated by both the demonstration of endogenous transcript regulation by miR-155-5p and validation of the miR-155-5p seed sequence through a reporter assay was recently assembled.[27] This list included 140 genes and included regulatory proteins for myelopoiesis and leukemogenesis (e.g. SHIP-1, AICDA, ETS1, JARID2, SPI1, etc.), inflammation (e.g. BACH1, FADD, IKBKE, INPP5D, MYD88, RIPK1, SPI1, SOCS, etc.) and known tumor suppressors (e.g. CEBPβ, IL17RB, PCCD4, TCF12, ZNF652, etc.).[27] The validated miR-155-5p binding site harbored in the SPI1 mRNA[28] and the validated miR-155-3p binding site harbored in the IRAK3 mRNA [25] are shown in Figures 4 and 5 respectively.


Physiological roles
Hematopoiesis
Hematopoiesis is defined as the formation and development of blood cells, all of which are derived from hematopoietic stem-progenitor cells (HSPCs).[29] HSPCs are primitive cells capable of self-renewal and initially differentiate into common myeloid progenitor (CMP) or common lymphoid progenitor (CLP) cells.[29] CMPs represent the cellular population that has become myeloid lineage and it is the point that myelopoiesis begins.[29] During myelopoiesis further cellular differentiation takes place including thrombopoiesis, erythropoiesis, granulopoiesis, and monocytopoiesis.[29] CLPs subsequently differentiate into B-cells and T-cells in a process designated lymphopoiesis.[29] Given that miR-155-5p is expressed in hematopoietic cells[22] it was hypothesized that this miRNA plays a critical role in these cellular differentiation processes. In support of this premise, miR-155-5p was found to be expressed in CD34(+) human HSPCs, and it was speculated that this miRNA may hold these cells at an early stem-progenitor stage, inhibiting their differentiation into a more mature cell (i.e. megakaryocytic/erythroid/granulocytic/monocytic/B-lymphoid/T-lymphoid).[30] This hypothesis was substantiated when pre-mir-155 transduced HSPCs generated 5-fold fewer myeloid and 3-fold fewer erythroid colonies.[30] Additionally, Hu et al.[31] demonstrated that the homeobox protein, HOXA9, regulated MIR155HG expression in myeloid cells and that this miRNA played a functional role in hematopoiesis. These investigators found that forced expression of miR-155-5p in bone marrow cells resulted in a ~50% decrease in SPI1 (i.e. PU.1),[31] a transcription factor and a regulator of myelopoiesis,[32] and a validated target of this miRNA.[28] It was also established that in vitro differentiation of purified human erythroid progenitor cells resulted in a progressive decrease of miR-155-5p expression in mature red cells.[33] Additionally, mice deficient in pre-mir-155 showed clear defects in lymphocyte development and generation of B- and T-cell responses in vivo.[28][34][35] Finally, it was established that regulatory T-cell (Tregs) development required miR-155-5p and this miRNA was shown to play a role in Treg homeostasis and overall survival by directly targeting SOCS1, a negative regulator for IL-2 signaling.[36][37] Taken together, these results strongly suggest that miR-155-5p is an essential molecule in the control of several aspects of hematopoiesis including myelopoiesis, erythropoiesis, and lymphopoiesis.
Immune system
The innate immune system constitutes the first line of defense against invading pathogens and is regarded as the major initiator of inflammatory responses.[38] Its cellular component involves primarily monocyte/macrophages, granulocytes, and dendritic cells (DCs), which are activated upon sensing of conserved pathogen structures (PAMPs) by pattern recognition receptors such as Toll-like receptors ((TLRs)).[39] MIR155HG (i.e. miR-155-5p) expression is greatly enhanced by TLR agonist stimulation of macrophages and dendritic cells.[40][41][42][43][44][45] Since microbial lipopolysaccharide (an agonist of TLR4) activates a chain of events that lead to the stimulation of the NF-κB and AP-1 transcription factors,[39] it was hypothesized that endotoxin activation of MIR155HG may be mediated by those transcription factors.[40] Indeed, MIR155HG expression was found to be activated in LPS treated murine macrophage cells (i.e. Raw264.7) by an NF-κB-mediated mechanism.[41] Furthermore, H. pylori infection of primary murine bone marrow-derived macrophages resulted in a NF-κB dependent up-regulation of MIR155HG.[46] In the context of viral infection vesicular stomatitis virus (VSV) challenge of murine peritoneal macrophages was reported to result in miR-155-5p over-expression via a retinoic acid-inducible gene I/JNK/NF-κB–dependent pathway.[47] Support for a role of AP-1 in MIR155HG activation comes from studies using stimuli relevant to viral infection such as TLR3 ligand poly(I:C) or interferon beta (IFN-β).[42] Downstream of those stimuli AP-1 seems to play a major role in MIR155HG activation.[42][48][49][50]
Upon its initiation via activation of e.g. TLRs by pathogen stimuli miR-155-5p functions as a post-transcriptional regulator of innate immune signaling pathways. Importantly, miR-155-5p displays a similar responsiveness to pathogen stimuli (e.g. TLR4 agonist LPS) as major pro-inflammatory marker mRNAs.[51] Once activated, miR-155-5p suppresses negative regulators of inflammation. These include inositol polyphosphate-5-phosphatase (INPP5D also denoted SHIP1) and suppressor of cytokine signaling 1 (SOCS1), suppression of which promotes cell survival, growth, migration, and anti-pathogen responses.[47][52][53][54] Besides supporting the activation of defense pathways miR-155-5p may also limit the strength of the resulting NF-κB dependent inflammatory response,[51] suggesting varying functions of miR-155 at different stages of inflammation.
Taken together, these observations imply that the activation of the MIR155HG may be context-dependent given that both AP-1- and NF-κB-mediated mechanisms regulate the expression of this gene. These studies also suggest that a broad range of viral and bacterial inflammatory mediators can stimulate the expression of miR-155-5p and indicate that there is an intimate relationship between inflammation, innate immunity and MIR155HG expression.
Activity and phenotypes
There is evidence that miR-155 participates in cascades associated with cardiovascular diseases and hypertension, and was also found to be implicated in immunity, genomic instability, cell differentiation, inflammation, virus associated infections, cancer, and diabetes mellitus.[55]
Protective roles of miR-155 may arise in response to its action on silencing genes thereby regulating their expression time, mutations in miR-155 target site deny it the optimal access necessary to bring about gene silencing, leading to over abundance of delinquent activities that may go malignant, for example, miR-155 role as a protective agent against predisposition to B Cell associated malignancies is emphasized by maintaining the balance of Activation-Induced Cytidine Deaminase (AID) enzyme. MiR-155 mediates regulation of AID abundance and expression time upon immunological cues however, mutations in the target on AID mRNA result in its unresponsiveness to miR-155 silencing and lead to unbridled expression of its protein causing wild immature B-lymphocyte surges and AID-mediated chromosomal translocations.[3][4]
Clinical significance
Cardiovascular
Transfection of miR-155 into human primary lung fibroblasts reduces the endogenous expression of the angiotensin II receptor AT1R protein. Furthermore, AT1R mediates angiotensin II-related elevation in blood pressure and contributes to the pathogenesis of heart failure. Defective miR-155 function could be implicated in hypertension and cardiovascular diseases if the cis-regulatory site on 3` UTR of AT1R (miR-155 target site) was affected due to a SNP polymorphism in AT1R itself. This mutation is disruptive of miR-155 targeting and thus preventive of AT1R expression down-regulation.[3] In low blood pressure over-expression of miR-155 correlates with the impairment of AT1R activity.[2]
Immunity
miR-155 is involved in immunity by playing key roles in modulating humoral and innate cell-mediated immune responses, for example, In miR-155 deficient mice, immunological-memory is impaired; making it fall prey to repetitive bouts of invasions by the same pathogen (Rodriguez et al. 2007), maturation and specificity of miR-155-deficient B-lymphocytes are impaired since the process relies on AID enzyme which has a miR-155 target in its 3′ UTR end.[3][4] The phenotypic consequences involving deficiency of miR-155 in mice show later in life where the animals develop lung and intestinal lesions.[2]
Activated B and T cells show increased miR-155 expression, the same goes for macrophages and dendritic cells of the immune system. MiR-155 is crucial for proper lymphocyte development and maturation. Details of various manifestations of miR-155 levels and involvement in activities that ascertain optimal immune responses have been the subject of many researches:
Reduction of IgG1
Defective T and B cells as well as markedly decreased IgG1 responses were observed in miR-155-deficient mice, IgG1 is reduced whereas the expression of the IgM immunoglobulin remains normal in these mice. The change in IgG1 levels maybe explained by the fact that it is a target for miR-155 in B cells, the protein-encoding mRNA for the transcriptional regulator Pu.1-protein, elevation of Pu.1 protein predisposes defective IgG1 production. In addition to Pu.1, there are nearly 60 other differentially elevated genes in miR-155 deficient B cells, further inspection revealed possible miR-155 target sites in the 3′ UTR regions in these genes.[4]
Lymphocyte malignancies
Mature receptors affinity and specificity of lymphocytes to pathogenic agents underlie proper immune responses, optimal miR-155 coordination is required for manufacturing of normal B lymphocytes, production of high-affinity antibodies and balancing of BCR signalling. It has been demonstrated that miR-155 can be transferred through gap junctions from leukemic cells to healthy B cells and promote their transformation to tumorigenic-like cells [56]
Selection of competent B cells takes place in the germinal center where they are trained to differentiate body cells vs. foreign antigens, they compete for antigen recognition and for T cell help, in this fashion of selective pressure those B Cells that demonstrated high-affinity receptors and cooperation with T cells (affinity maturation) are recruited and deployed to the bone marrow or become memory B cells, apoptotic termination takes place for those B Cells failing the competition. Immature B cells which are miR-155 deficient evade apoptosis as a result of elevated Bcl-2 protein levels; a protein that was found to be involved in B Cell malignancies and to be controlled by miR-155.[4]
Inflammation
Inflammatory responses to triggers such as TNF-α involve macrophages with components that include miR-155. miR-155 is overexpressed in atopic dermatitis and contributes to chronic skin inflammation by increasing the proliferative response of T(H) cells through the downregulation of CTLA-4.[57] In Autoimmune disorders such as rheumatoid arthritis, miR-155 showed higher expression in patients' tissues and synovial fibroblasts.[2] In multiple sclerosis, increased expression of mir-155 has also been measured in peripheral and CNS-resident myeloid cells, including circulating blood monocytes and activated microglia.[58] It was also found that mir-155 is implicated in inflammation. Overexpression of mir-155 will lead to chronic inflammatory state in human.[59]
DNA viruses
In DNA viruses, miRNAs were experimentally verified, miRNAs in viruses are encoded by dsDNAs,[3] examples of such viruses include herpesviruses such as Humans-Epstein-Barr Virus (EBV) and adenoviruses,[2] another virus expressing miR-155-like miRNA in chickens is the oncogenic MDV-1 whose non-oncogenic relative MDV-2 does not, this suggests implication of miR-155 in lymphomagenesis.[3] Viruses can exploit host miRNAs to the degree that they use host miRNAs to encode for viral clones for example: miR-K12-11 in Kaposi's-sarcoma-associated Herpesvirus has a target specificity region orthologous to that of miR-155's; mimicking the action of miR-155 [60] and, sharing targets with it, thus it can be thought to suppress miR-155 accessibility to its targets by competition and this in effect downregulates expression of genes playing roles in cellular growth and apoptosis in a manner that defies regulations by miR-155.[2] EBV modulates host miR-155 expression, which is essential for growth of EBV-infected B cells.[61] EBV-infected cells have increased expression of miR-155 thereby disturbing equilibrium of expression for genes regulating transcription in those cells.[2][3]
Cancer
Over-silencing by miR-155 may result in triggering oncogenic cascades that begin by apoptotic resistance, the pro-apoptotic Tumour Protein-53-induced-nuclear-protein1 (TP53INP1) is silenced by miR-155, over-expression of miR-155 leads to decreased levels of TP53INP1 in pancreatic ductal adenocarcinomas and possibly in other epithelial cancers where TP53INP1 activity is lost thereby resulting in apoptosis evasion and uncontrolled bouts of growth.[3]
Inactivation of DNA Mismatch Repair (MMR) as identified by elevation of mutation rates is the cause of Lynch Syndrome (LS), also known as hereditary nonpolyposis colorectal cancer (HNPCC), down-regulation of MMR controlling protein is carried out by over-expression of miR-155, MMR is controlled by a group of conserved proteins, reduced activity of these proteins results in elevated levels of mutations in the phenotype triggering a march towards developing this type of cancer.[62]
Other types of tumors in which miR-155 over-expression was reported include: thyroid carcinoma, breast cancer, colon cancer, cervical cancer, and lung cancer, where distinct miR-155 expression profiles quantification can potentially serve as signals for tumor detection and evaluation of prognosis outcome.[2] It is shown in an analysis that miR-155 expression is associated with survival in triple negative breast cancer.[63]
Notes
See also
References
- ↑ "Entrez Gene: MIR155HG". https://www.ncbi.nlm.nih.gov/gene/114614.
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 2.8 "miR-155 gene: a typical multifunctional microRNA". Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 1792 (6): 497–505. Jun 2009. doi:10.1016/j.bbadis.2009.02.013. PMID 19268705. https://hal.archives-ouvertes.fr/hal-00488806/file/PEER_stage2_10.1016%252Fj.bbadis.2009.02.013.pdf.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 3.8 "Shhh! Silencing by microRNA-155". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences 364 (1517): 631–637. Mar 2009. doi:10.1098/rstb.2008.0209. PMID 19008191.
- ↑ 4.0 4.1 4.2 4.3 4.4 4.5 "MicroRNA-155 function in B Cells". Immunity 27 (6): 825–827. Dec 2007. doi:10.1016/j.immuni.2007.11.010. PMID 18093533.
- ↑ "miR-155: on the crosstalk between inflammation and cancer". International Reviews of Immunology 28 (5): 264–284. 2009. doi:10.1080/08830180903093796. PMID 19811312.
- ↑ "microRNA regulation of inflammatory responses". Annual Review of Immunology 30: 295–312. 2012. doi:10.1146/annurev-immunol-020711-075013. PMID 22224773.
- ↑ "Regulation of the MIR155 host gene in physiological and pathological processes". Gene 532 (1): 1–12. Dec 2013. doi:10.1016/j.gene.2012.12.009. PMID 23246696.
- ↑ "The oncogenic role of miR-155 in breast cancer". Cancer Epidemiology, Biomarkers & Prevention 21 (8): 1236–1243. Aug 2012. doi:10.1158/1055-9965.EPI-12-0173. PMID 22736789.
- ↑ "Nanoparticle-based therapy in an in vivo microRNA-155 (miR-155)-dependent mouse model of lymphoma". Proceedings of the National Academy of Sciences of the United States of America 109 (26): E1695–1704. Jun 2012. doi:10.1073/pnas.1201516109. PMID 22685206.
- ↑ "Oncogenic IRFs provide a survival advantage for Epstein-Barr virus- or human T-cell leukemia virus type 1-transformed cells through induction of BIC expression". Journal of Virology 85 (16): 8328–8337. Aug 2011. doi:10.1128/JVI.00570-11. PMID 21680528.
- ↑ "MicroRNA profiling identifies microRNA-155 as an adverse mediator of cardiac injury and dysfunction during acute viral myocarditis". Circulation Research 111 (4): 415–425. Aug 2012. doi:10.1161/CIRCRESAHA.112.267443. PMID 22715471.
- ↑ "Accumulation of miR-155 and BIC RNA in human B cell lymphomas". Proceedings of the National Academy of Sciences of the United States of America 102 (10): 3627–3632. Mar 2005. doi:10.1073/pnas.0500613102. PMID 15738415. Bibcode: 2005PNAS..102.3627E.
- ↑ 13.0 13.1 13.2 13.3 "Identification and characterization of human BIC, a gene on chromosome 21 that encodes a noncoding RNA". Gene 274 (1–2): 157–167. Aug 2001. doi:10.1016/S0378-1119(01)00612-6. PMID 11675008.
- ↑ 14.0 14.1 "Biogenesis of small RNAs in animals". Nature Reviews Molecular Cell Biology 10 (2): 126–139. Feb 2009. doi:10.1038/nrm2632. PMID 19165215.
- ↑ 15.0 15.1 "The widespread regulation of microRNA biogenesis, function and decay". Nature Reviews Genetics 11 (9): 597–610. Sep 2010. doi:10.1038/nrg2843. PMID 20661255.
- ↑ 16.0 16.1 16.2 "The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC". Nature Structural & Molecular Biology 19 (6): 586–593. Jun 2012. doi:10.1038/nsmb.2296. PMID 22664986.
- ↑ "microRNA functions". Annual Review of Cell and Developmental Biology 23: 175–205. 2007. doi:10.1146/annurev.cellbio.23.090506.123406. PMID 17506695.
- ↑ "Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?". Nature Reviews Genetics 9 (2): 102–114. Feb 2008. doi:10.1038/nrg2290. PMID 18197166. https://escholarship.mcgill.ca/concern/articles/cz30pz55f.
- ↑ "The microRNA Registry". Nucleic Acids Research 32 (Database issue): D109–11. Jan 2004. doi:10.1093/nar/gkh023. PMID 14681370.
- ↑ 20.0 20.1 "Most mammalian mRNAs are conserved targets of microRNAs". Genome Research 19 (1): 92–105. Jan 2009. doi:10.1101/gr.082701.108. PMID 18955434.
- ↑ "MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts". The Journal of Biological Chemistry 281 (27): 18277–18284. Jul 2006. doi:10.1074/jbc.M601496200. PMID 16675453.
- ↑ 22.0 22.1 22.2 "A mammalian microRNA expression atlas based on small RNA library sequencing". Cell 129 (7): 1401–1414. Jun 2007. doi:10.1016/j.cell.2007.04.040. PMID 17604727.
- ↑ "Trisomy-21 gene dosage over-expression of miRNAs results in the haploinsufficiency of specific target proteins". RNA Biology 7 (5): 540–547. 2010. doi:10.4161/rna.7.5.12685. PMID 21081842.
- ↑ "Interferon regulatory factor 3 inhibits astrocyte inflammatory gene expression through suppression of the proinflammatory miR-155 and miR-155*". Glia 59 (12): 1911–1922. Dec 2011. doi:10.1002/glia.21233. PMID 22170100.
- ↑ 25.0 25.1 "miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells". Blood 116 (26): 5885–5894. Dec 2010. doi:10.1182/blood-2010-04-280156. PMID 20852130.
- ↑ "Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets". Cell 120 (1): 15–20. Jan 2005. doi:10.1016/j.cell.2004.12.035. PMID 15652477.
- ↑ 27.0 27.1 "Mutant p53 drives invasion in breast tumors through up-regulation of miR-155". Oncogene 32 (24): 2992–3000. Jun 2013. doi:10.1038/onc.2012.305. PMID 22797073.
- ↑ 28.0 28.1 28.2 "microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells". Immunity 27 (6): 847–859. Dec 2007. doi:10.1016/j.immuni.2007.10.009. PMID 18055230.
- ↑ 29.0 29.1 29.2 29.3 29.4 "Biological differences between neonatal and adult human hematopoietic stem/progenitor cells". Stem Cells and Development 19 (3): 285–298. Mar 2010. doi:10.1089/scd.2009.0327. PMID 19778207.
- ↑ 30.0 30.1 "CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control". Proceedings of the National Academy of Sciences of the United States of America 104 (8): 2750–2755. Feb 2007. doi:10.1073/pnas.0610983104. PMID 17293455. Bibcode: 2007PNAS..104.2750G.
- ↑ 31.0 31.1 "HOXA9 regulates miR-155 in hematopoietic cells". Nucleic Acids Research 38 (16): 5472–5478. Sep 2010. doi:10.1093/nar/gkq337. PMID 20444872.
- ↑ "PU.1: a crucial and versatile player in hematopoiesis and leukemia". The International Journal of Biochemistry & Cell Biology 40 (1): 22–27. 2008. doi:10.1016/j.biocel.2007.01.026. PMID 17374502.
- ↑ "Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis". Biochemical and Biophysical Research Communications 364 (3): 509–514. Dec 2007. doi:10.1016/j.bbrc.2007.10.077. PMID 17964546.
- ↑ "Requirement of bic/microRNA-155 for normal immune function". Science 316 (5824): 608–611. Apr 2007. doi:10.1126/science.1139253. PMID 17463290. Bibcode: 2007Sci...316..608R.
- ↑ "Regulation of the germinal center response by microRNA-155". Science 316 (5824): 604–608. Apr 2007. doi:10.1126/science.1141229. PMID 17463289. Bibcode: 2007Sci...316..604T.
- ↑ "Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells". Journal of Immunology 182 (5): 2578–2582. Mar 2009. doi:10.4049/jimmunol.0803162. PMID 19234151.
- ↑ "Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein". Immunity 30 (1): 80–91. Jan 2009. doi:10.1016/j.immuni.2008.11.010. PMID 19144316.
- ↑ "Inflammation 2010: new adventures of an old flame". Cell 140 (6): 771–776. Mar 2010. doi:10.1016/j.cell.2010.03.006. PMID 20303867.
- ↑ 39.0 39.1 "Toll-like receptors in innate immunity". International Immunology 17 (1): 1–14. Jan 2005. doi:10.1093/intimm/dxh186. PMID 15585605.
- ↑ 40.0 40.1 "NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses". Proceedings of the National Academy of Sciences of the United States of America 103 (33): 12481–12486. Aug 2006. doi:10.1073/pnas.0605298103. PMID 16885212. Bibcode: 2006PNAS..10312481T.
- ↑ 41.0 41.1 "Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock". Journal of Immunology 179 (8): 5082–5089. Oct 2007. doi:10.4049/jimmunol.179.8.5082. PMID 17911593.
- ↑ 42.0 42.1 42.2 "MicroRNA-155 is induced during the macrophage inflammatory response". Proceedings of the National Academy of Sciences of the United States of America 104 (5): 1604–1609. Jan 2007. doi:10.1073/pnas.0610731104. PMID 17242365. Bibcode: 2007PNAS..104.1604O.
- ↑ "MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells". Proceedings of the National Academy of Sciences of the United States of America 106 (8): 2735–2740. Feb 2009. doi:10.1073/pnas.0811073106. PMID 19193853. Bibcode: 2009PNAS..106.2735C.
- ↑ "MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response". PLOS ONE 4 (12): e8508. 2009. doi:10.1371/journal.pone.0008508. PMID 20041145. Bibcode: 2009PLoSO...4.8508C.
- ↑ "In vivo microRNA-155 expression influences antigen-specific T cell-mediated immune responses generated by DNA vaccination". Cell & Bioscience 1 (1): 3. 2011. doi:10.1186/2045-3701-1-3. PMID 21711593.
- ↑ "Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis". Proceedings of the National Academy of Sciences of the United States of America 109 (19): E1153–1162. May 2012. doi:10.1073/pnas.1116125109. PMID 22509021.
- ↑ 47.0 47.1 "Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1". Journal of Immunology 185 (10): 6226–6233. Nov 2010. doi:10.4049/jimmunol.1000491. PMID 20937844.
- ↑ "Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD". Carcinogenesis 31 (9): 1561–1566. Sep 2010. doi:10.1093/carcin/bgq143. PMID 20622002.
- ↑ "IL-10 inhibits miR-155 induction by toll-like receptors". The Journal of Biological Chemistry 285 (27): 20492–20498. Jul 2010. doi:10.1074/jbc.M110.102111. PMID 20435894.
- ↑ "LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages". FASEB Journal 23 (9): 2898–2908. Sep 2009. doi:10.1096/fj.09-131342. PMID 19423639.
- ↑ 51.0 51.1 "Differential activation and functional specialization of miR-146 and miR-155 in innate immune sensing". Nucleic Acids Research 41 (1): 542–553. Jan 2013. doi:10.1093/nar/gks1030. PMID 23143100.
- ↑ "The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs". Immunity 31 (2): 220–231. Aug 2009. doi:10.1016/j.immuni.2009.06.024. PMID 19699171.
- ↑ "Inositol phosphatase SHIP1 is a primary target of miR-155". Proceedings of the National Academy of Sciences of the United States of America 106 (17): 7113–7118. Apr 2009. doi:10.1073/pnas.0902636106. PMID 19359473. Bibcode: 2009PNAS..106.7113O.
- ↑ "Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice". Blood 114 (7): 1374–1382. Aug 2009. doi:10.1182/blood-2009-05-220814. PMID 19520806.
- ↑ "Functional Role of miR-155 in the Pathogenesis of Diabetes Mellitus and Its Complications". Non-Coding RNA 7 (3): 39. July 2021. doi:10.3390/ncrna7030039. PMID 34287359.
- ↑ "mIRNA-155 shuttling through gap junctions facilitates CLL progression". The 14th CIMT Annual Meeting Mechanisms of Efficacy in Cancer Immunotherapy, Mainz, Germany, 2016. 2016.
- ↑ "MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4". The Journal of Allergy and Clinical Immunology 126 (3): 581–589.e1–20. Sep 2010. doi:10.1016/j.jaci.2010.05.045. PMID 20673989.
- ↑ "miR-155 as a multiple sclerosis-relevant regulator of myeloid cell polarization". Annals of Neurology 74 (5): 709–720. Nov 2013. doi:10.1002/ana.23967. PMID 23818336.
- ↑ "microRNA regulation of inflammatory responses". Annual Review of Immunology 30: 295–312. 2012. doi:10.1146/annurev-immunol-020711-075013. PMID 22224773.
- ↑ "Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155". Journal of Virology 81 (23): 12836–12845. Dec 2007. doi:10.1128/JVI.01804-07. PMID 17881434.
- ↑ "Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus". Journal of Virology 84 (22): 11670–11678. Nov 2010. doi:10.1128/JVI.01248-10. PMID 20844043.
- ↑ "Modulation of mismatch repair and genomic stability by miR-155". Proceedings of the National Academy of Sciences of the United States of America 107 (15): 6982–6987. Apr 2010. doi:10.1073/pnas.1002472107. PMID 20351277. Bibcode: 2010PNAS..107.6982V.
- ↑ Lánczky, András; Nagy, Ádám; Bottai, Giulia; Munkácsy, Gyöngyi; Szabó, András; Santarpia, Libero; Győrffy, Balázs (2016-12-01). "miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients". Breast Cancer Research and Treatment 160 (3): 439–446. doi:10.1007/s10549-016-4013-7. ISSN 1573-7217. PMID 27744485.
Further reading
- "Identification of hundreds of conserved and nonconserved human microRNAs". Nature Genetics 37 (7): 766–770. Jul 2005. doi:10.1038/ng1590. PMID 15965474.
- "MicroRNA-155 silencing enhances inflammatory response and lipid uptake in oxidized low-density lipoprotein-stimulated human THP-1 macrophages". Journal of Investigative Medicine 58 (8): 961–967. Dec 2010. doi:10.2310/jim.0b013e3181ff46d7. PMID 21030878.
- "Interplay between miR-155, AT1R A1166C polymorphism, and AT1R expression in young untreated hypertensives". American Journal of Hypertension 24 (2): 241–246. Feb 2011. doi:10.1038/ajh.2010.211. PMID 20966899.
- "Serum and urinary cell-free MiR-146a and MiR-155 in patients with systemic lupus erythematosus". The Journal of Rheumatology 37 (12): 2516–2522. Dec 2010. doi:10.3899/jrheum.100308. PMID 20952466.
- "Inflammatory cytokines regulate microRNA-155 expression in human retinal pigment epithelial cells by activating JAK/STAT pathway". Biochemical and Biophysical Research Communications 402 (2): 390–395. Nov 2010. doi:10.1016/j.bbrc.2010.10.042. PMID 20950585.
- "IFN-γ and TNF-α synergistically induce microRNA-155 which regulates TAB2/IP-10 expression in human mesangial cells". American Journal of Nephrology 32 (5): 462–468. 2010. doi:10.1159/000321365. PMID 20948191.
- "NF-kappaB down-regulates expression of the B-lymphoma marker CD10 through a miR-155/PU.1 pathway". The Journal of Biological Chemistry 286 (3): 1675–1682. Jan 2011. doi:10.1074/jbc.M110.177063. PMID 20947507.
- "Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1". Journal of Immunology 185 (10): 6226–6233. Nov 2010. doi:10.4049/jimmunol.1000491. PMID 20937844.
- "MicroRNA-155 promotes autoimmune inflammation by enhancing inflammatory T cell development". Immunity 33 (4): 607–619. Oct 2010. doi:10.1016/j.immuni.2010.09.009. PMID 20888269.
- "Human SMG-1 is involved in gemcitabine-induced primary microRNA-155/BIC up-regulation in human pancreatic cancer PANC-1 cells". Pancreas 40 (1): 55–60. Jan 2011. doi:10.1097/MPA.0b013e3181e89f74. PMID 20871480.
- "miR-155 and its star-form partner miR-155* cooperatively regulate type I interferon production by human plasmacytoid dendritic cells". Blood 116 (26): 5885–5894. Dec 2010. doi:10.1182/blood-2010-04-280156. PMID 20852130.
- "Virally induced cellular microRNA miR-155 plays a key role in B-cell immortalization by Epstein-Barr virus". Journal of Virology 84 (22): 11670–11678. Nov 2010. doi:10.1128/JVI.01248-10. PMID 20844043.
- "MicroRNA-155 regulates angiotensin II type 1 receptor expression and phenotypic differentiation in vascular adventitial fibroblasts". Biochemical and Biophysical Research Communications 400 (4): 483–488. Oct 2010. doi:10.1016/j.bbrc.2010.08.067. PMID 20735984.
- "Resveratrol decreases the levels of miR-155 by upregulating miR-663, a microRNA targeting JunB and JunD". Carcinogenesis 31 (9): 1561–1566. Sep 2010. doi:10.1093/carcin/bgq143. PMID 20622002.
- "Effect of quercetin and its metabolites isorhamnetin and quercetin-3-glucuronide on inflammatory gene expression: role of miR-155". The Journal of Nutritional Biochemistry 22 (3): 293–299. Mar 2011. doi:10.1016/j.jnutbio.2010.02.008. PMID 20579867.
- "MicroRNA-155 prevents necrotic cell death in human cardiomyocyte progenitor cells via targeting RIP1". Journal of Cellular and Molecular Medicine 15 (7): 1474–1482. Jul 2011. doi:10.1111/j.1582-4934.2010.01104.x. PMID 20550618.
- "Difference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice". Laboratory Investigation 90 (10): 1437–1446. Oct 2010. doi:10.1038/labinvest.2010.113. PMID 20548288.
- "MicroRNA-155 contributes to preeclampsia by down-regulating CYR61". American Journal of Obstetrics and Gynecology 202 (5): 466.e1–7. May 2010. doi:10.1016/j.ajog.2010.01.057. PMID 20452491.
- "HOXA9 regulates miR-155 in hematopoietic cells". Nucleic Acids Research 38 (16): 5472–5478. Sep 2010. doi:10.1093/nar/gkq337. PMID 20444872.
- "IL-10 inhibits miR-155 induction by toll-like receptors". The Journal of Biological Chemistry 285 (27): 20492–20498. Jul 2010. doi:10.1074/jbc.M110.102111. PMID 20435894.
- "MicroRNA miR-155 inhibits bone morphogenetic protein (BMP) signaling and BMP-mediated Epstein-Barr virus reactivation". Journal of Virology 84 (13): 6318–6327. Jul 2010. doi:10.1128/JVI.00635-10. PMID 20427544.
- "[Expression and its clinical significance of miR-155 in human primary breast cancer]". Zhonghua Wai Ke Za Zhi [Chinese Journal of Surgery] 48 (3): 205–208. Feb 2010. PMID 20388420.
- "MicroRNA-155 regulates cell survival, growth, and chemosensitivity by targeting FOXO3a in breast cancer". The Journal of Biological Chemistry 285 (23): 17869–17879. Jun 2010. doi:10.1074/jbc.M110.101055. PMID 20371610.
- "[Effect of anti-cancer drugs on the expression of BIC/miR-155 in human pancreatic cancer PANC-1 cells]". Zhonghua Yi Xue Za Zhi 90 (2): 123–127. Jan 2010. PMID 20356498.
- "MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene". Cancer Research 70 (8): 3119–3127. Apr 2010. doi:10.1158/0008-5472.CAN-09-4250. PMID 20354188.
- "Aberrant MicroRNA-155 expression is an early event in the multistep progression of pancreatic adenocarcinoma". Pancreatology 10 (1): 66–73. 2010. doi:10.1159/000231984. PMID 20332664.
- "Efficient inhibition of miR-155 function in vivo by peptide nucleic acids". Nucleic Acids Research 38 (13): 4466–4475. Jul 2010. doi:10.1093/nar/gkq160. PMID 20223773.
- "Identification of MyD88 as a novel target of miR-155, involved in negative regulation of Helicobacter pylori-induced inflammation". FEBS Letters 584 (8): 1481–1486. Apr 2010. doi:10.1016/j.febslet.2010.02.063. PMID 20219467.
- "Helicobacter pylori induces miR-155 in T cells in a cAMP-Foxp3-dependent manner". PLOS ONE 5 (3): e9500. 2010. doi:10.1371/journal.pone.0009500. PMID 20209161. Bibcode: 2010PLoSO...5.9500F.
- "Expression of microRNA-155 precursor in peripheral blood mononuclear cells from Hepatitis C patients after antiviral treatment". Acta Virologica 54 (1): 75–78. 2010. doi:10.4149/av_2010_01_75. PMID 20201617.
- "Teratogen-induced alterations in microRNA-34, microRNA-125b and microRNA-155 expression: correlation with embryonic p53 genotype and limb phenotype". BMC Developmental Biology 10: 20. 2010. doi:10.1186/1471-213X-10-20. PMID 20170545.
- "Targeting of SMAD5 links microRNA-155 to the TGF-beta pathway and lymphomagenesis". Proceedings of the National Academy of Sciences of the United States of America 107 (7): 3111–3116. Feb 2010. doi:10.1073/pnas.0910667107. PMID 20133617. Bibcode: 2010PNAS..107.3111R.
- "MiR-155 induction by F. novicida but not the virulent F. tularensis results in SHIP down-regulation and enhanced pro-inflammatory cytokine response". PLOS ONE 4 (12): e8508. 2009. doi:10.1371/journal.pone.0008508. PMID 20041145. Bibcode: 2009PLoSO...4.8508C.
- "Induction of microRNAs, mir-155, mir-222, mir-424 and mir-503, promotes monocytic differentiation through combinatorial regulation". Leukemia 24 (2): 460–466. Feb 2010. doi:10.1038/leu.2009.246. PMID 19956200.
- "Onco-miR-155 targets SHIP1 to promote TNFalpha-dependent growth of B cell lymphomas". EMBO Molecular Medicine 1 (5): 288–295. Aug 2009. doi:10.1002/emmm.200900028. PMID 19890474.
- "MicroRNA-155 regulates inflammatory cytokine production in tumor-associated macrophages via targeting C/EBPbeta". Cellular & Molecular Immunology 6 (5): 343–352. Oct 2009. doi:10.1038/cmi.2009.45. PMID 19887047.
- "miR-155: on the crosstalk between inflammation and cancer". International Reviews of Immunology 28 (5): 264–284. 2009. doi:10.1080/08830180903093796. PMID 19811312.
- "miR-155 inhibition sensitizes CD4+ Th cells for TREG mediated suppression". PLOS ONE 4 (9): e7158. 2009. doi:10.1371/journal.pone.0007158. PMID 19777054. Bibcode: 2009PLoSO...4.7158S.
- "Reticuloendotheliosis virus strain T induces miR-155, which targets JARID2 and promotes cell survival". Journal of Virology 83 (23): 12009–12017. Dec 2009. doi:10.1128/JVI.01182-09. PMID 19759154.
- "Role of microRNA-155 at early stages of hepatocarcinogenesis induced by choline-deficient and amino acid-defined diet in C57BL/6 mice". Hepatology 50 (4): 1152–1161. Oct 2009. doi:10.1002/hep.23100. PMID 19711427.
- "Identification of keratinocyte growth factor as a target of microRNA-155 in lung fibroblasts: implication in epithelial-mesenchymal interactions". PLOS ONE 4 (8): e6718. 2009. doi:10.1371/journal.pone.0006718. PMID 19701459. Bibcode: 2009PLoSO...4.6718P.
- "Induction of microRNA-155 during Helicobacter pylori infection and its negative regulatory role in the inflammatory response". The Journal of Infectious Diseases 200 (6): 916–925. Sep 2009. doi:10.1086/605443. PMID 19650740.
- "Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF". Nucleic Acids Research 37 (17): 5784–5792. Sep 2009. doi:10.1093/nar/gkp577. PMID 19596814.
- "Src homology 2 domain-containing inositol-5-phosphatase and CCAAT enhancer-binding protein beta are targeted by miR-155 in B cells of Emicro-MiR-155 transgenic mice". Blood 114 (7): 1374–1382. Aug 2009. doi:10.1182/blood-2009-05-220814. PMID 19520806.
- "LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages". FASEB Journal 23 (9): 2898–2908. Sep 2009. doi:10.1096/fj.09-131342. PMID 19423639.
- "MicroRNA-155 modulates the pathogen binding ability of dendritic cells (DCs) by down-regulation of DC-specific intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN)". The Journal of Biological Chemistry 284 (24): 16334–16342. Jun 2009. doi:10.1074/jbc.M109.011601. PMID 19386588.
- "Inositol phosphatase SHIP1 is a primary target of miR-155". Proceedings of the National Academy of Sciences of the United States of America 106 (17): 7113–7118. Apr 2009. doi:10.1073/pnas.0902636106. PMID 19359473. Bibcode: 2009PNAS..106.7113O.
- "Cutting edge: the Foxp3 target miR-155 contributes to the development of regulatory T cells". Journal of Immunology 182 (5): 2578–2582. Mar 2009. doi:10.4049/jimmunol.0803162. PMID 19234151.
- "MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells". Proceedings of the National Academy of Sciences of the United States of America 106 (8): 2735–2740. Feb 2009. doi:10.1073/pnas.0811073106. PMID 19193853. Bibcode: 2009PNAS..106.2735C.
- "MicroRNA miR-155 is a biomarker of early pancreatic neoplasia". Cancer Biology & Therapy 8 (4): 340–346. Feb 2009. doi:10.4161/cbt.8.4.7338. PMID 19106647.
- "MicroRNA-155 expression and outcome in diffuse large B-cell lymphoma". British Journal of Haematology 144 (1): 138–140. Jan 2009. doi:10.1111/j.1365-2141.2008.07424.x. PMID 19016736.
- "MicroRNA 155 modulates megakaryopoiesis at progenitor and precursor level by targeting Ets-1 and Meis1 transcription factors". British Journal of Haematology 143 (4): 570–580. Nov 2008. doi:10.1111/j.1365-2141.2008.07382.x. PMID 18950466.
- "A functional MicroRNA-155 ortholog encoded by the oncogenic Marek's disease virus". Journal of Virology 83 (1): 489–492. Jan 2009. doi:10.1128/JVI.01166-08. PMID 18945769.
- "Epstein-Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NF-kappaB pathway". Nucleic Acids Research 36 (20): 6608–6619. Nov 2008. doi:10.1093/nar/gkn666. PMID 18940871.
- "Latent membrane protein-1 of Epstein-Barr virus induces the expression of B-cell integration cluster, a precursor form of microRNA-155, in B lymphoma cell lines". Biochemical and Biophysical Research Communications 377 (2): 579–583. Dec 2008. doi:10.1016/j.bbrc.2008.10.007. PMID 18926796.
- "MicroRNA-155 is regulated by the transforming growth factor beta/Smad pathway and contributes to epithelial cell plasticity by targeting RhoA". Molecular and Cellular Biology 28 (22): 6773–6784. Nov 2008. doi:10.1128/MCB.00941-08. PMID 18794355.
- "Epstein-Barr virus-induced miR-155 attenuates NF-kappaB signaling and stabilizes latent virus persistence". Journal of Virology 82 (21): 10436–10443. Nov 2008. doi:10.1128/JVI.00752-08. PMID 18753206.
- "MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation". Immunity 28 (5): 630–638. May 2008. doi:10.1016/j.immuni.2008.04.002. PMID 18455451.
- "MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase". Immunity 28 (5): 621–629. May 2008. doi:10.1016/j.immuni.2008.03.015. PMID 18450484.
- "MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways". Journal of Virology 82 (11): 5295–5306. Jun 2008. doi:10.1128/JVI.02380-07. PMID 18367535.
- "BIC is processed efficiently to microRNA-155 in Burkitt lymphoma cells". Leukemia 22 (9): 1795–1797. Sep 2008. doi:10.1038/leu.2008.62. PMID 18354490.
- "miRNA analysis in B-cell chronic lymphocytic leukaemia: proliferation centres characterized by low miR-150 and high BIC/miR-155 expression". The Journal of Pathology 215 (1): 13–20. May 2008. doi:10.1002/path.2333. PMID 18348159.
- "Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder". The Journal of Experimental Medicine 205 (3): 585–594. Mar 2008. doi:10.1084/jem.20072108. PMID 18299402.
- "Coordinated expression of microRNA-155 and predicted target genes in diffuse large B-cell lymphoma". Cancer Genetics and Cytogenetics 181 (1): 8–15. Feb 2008. doi:10.1016/j.cancergencyto.2007.10.008. PMID 18262046.
- "A viral microRNA functions as an orthologue of cellular miR-155". Nature 450 (7172): 1096–1099. Dec 2007. doi:10.1038/nature05992. PMID 18075594. Bibcode: 2007Natur.450.1096G.
- "microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells". Immunity 27 (6): 847–859. Dec 2007. doi:10.1016/j.immuni.2007.10.009. PMID 18055230.
- "B-cell receptor activation induces BIC/miR-155 expression through a conserved AP-1 element". The Journal of Biological Chemistry 283 (5): 2654–2662. Feb 2008. doi:10.1074/jbc.M708218200. PMID 18048365.
- "Modulation of miR-155 and miR-125b levels following lipopolysaccharide/TNF-alpha stimulation and their possible roles in regulating the response to endotoxin shock". Journal of Immunology 179 (8): 5082–5089. Oct 2007. doi:10.4049/jimmunol.179.8.5082. PMID 17911593.
- "Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development". Proceedings of the National Academy of Sciences of the United States of America 104 (41): 16170–16175. Oct 2007. doi:10.1073/pnas.0703942104. PMID 17911264. Bibcode: 2007PNAS..10416170G.
- "Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155". Journal of Virology 81 (23): 12836–12845. Dec 2007. doi:10.1128/JVI.01804-07. PMID 17881434.
- "Human microRNA-155 on chromosome 21 differentially interacts with its polymorphic target in the AGTR1 3′ untranslated region: a mechanism for functional single-nucleotide polymorphisms related to phenotypes". American Journal of Human Genetics 81 (2): 405–413. Aug 2007. doi:10.1086/519979. PMID 17668390.
- "The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microRNA-155 binding". The Journal of Biological Chemistry 282 (33): 24262–24269. Aug 2007. doi:10.1074/jbc.M701050200. PMID 17588946.
- "Requirement of bic/microRNA-155 for normal immune function". Science 316 (5824): 608–611. Apr 2007. doi:10.1126/science.1139253. PMID 17463290. Bibcode: 2007Sci...316..608R.
- "Regulation of the germinal center response by microRNA-155". Science 316 (5824): 604–608. Apr 2007. doi:10.1126/science.1141229. PMID 17463289. Bibcode: 2007Sci...316..604T.
- "MicroRNA-155 is induced during the macrophage inflammatory response". Proceedings of the National Academy of Sciences of the United States of America 104 (5): 1604–1609. Jan 2007. doi:10.1073/pnas.0610731104. PMID 17242365. Bibcode: 2007PNAS..104.1604O.
- "MicroRNA-155 regulates human angiotensin II type 1 receptor expression in fibroblasts". The Journal of Biological Chemistry 281 (27): 18277–18284. Jul 2006. doi:10.1074/jbc.M601496200. PMID 16675453.
- "Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155". Nucleic Acids Research 34 (7): e53. 2006. doi:10.1093/nar/gkl143. PMID 16614444.
- "miR-155/BIC as an oncogenic microRNA". Genes, Chromosomes & Cancer 45 (2): 211–212. Feb 2006. doi:10.1002/gcc.20282. PMID 16252262.
- "Lack of BIC and microRNA miR-155 expression in primary cases of Burkitt lymphoma". Genes, Chromosomes & Cancer 45 (2): 147–153. Feb 2006. doi:10.1002/gcc.20273. PMID 16235244.
- "Increased expression of microRNA-155 in Epstein-Barr virus transformed lymphoblastoid cell lines". Genes, Chromosomes & Cancer 45 (1): 103–106. Jan 2006. doi:10.1002/gcc.20264. PMID 16175574.
- "BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas". The Journal of Pathology 207 (2): 243–249. Oct 2005. doi:10.1002/path.1825. PMID 16041695.
- "Accumulation of miR-155 and BIC RNA in human B cell lymphomas". Proceedings of the National Academy of Sciences of the United States of America 102 (10): 3627–3632. Mar 2005. doi:10.1073/pnas.0500613102. PMID 15738415. Bibcode: 2005PNAS..102.3627E.
- "High expression of precursor microRNA-155/BIC RNA in children with Burkitt lymphoma". Genes, Chromosomes & Cancer 39 (2): 167–169. Feb 2004. doi:10.1002/gcc.10316. PMID 14695998.
- Xu, Yanan; Wu, Junhua; Yuan, Xiaoqi; Liu, Wenyuan; Pan, Jiewen; Xu, Binbin (2021). "MicroRNA-155 contributes to host immunity against Toxoplasma gondii". Parasite 28: 83. doi:10.1051/parasite/2021082. PMID 34907898.

External links
 |

