Chemistry:CP-532,903
From HandWiki
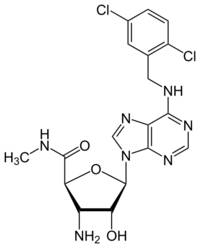
| |
| Names | |
|---|---|
| IUPAC name
6-{[(2,5-Dichlorophenyl)methyl]amino}-9H-purin-9-yl 3-amino-3-deoxy-N-methyl-β-D-ribofuranosiduronamide
| |
| Systematic IUPAC name
(2S,3S,4R,5R)-3-Amino-5-(6-{[(2,5-dichlorophenyl)methyl]amino}-9H-purin-9-yl)-4-hydroxy-N5-methyloxolane-2-carboxamide | |
| Other names
CP-532,903
| |
| Identifiers | |
3D model (JSmol)
|
|
| ChemSpider | |
PubChem CID
|
|
| UNII | |
| |
| |
| Properties | |
| C18H19Cl2N7O3 | |
| Molar mass | 452.294 |
Except where otherwise noted, data are given for materials in their standard state (at 25 °C [77 °F], 100 kPa). | |
| Infobox references | |
Tracking categories (test):
CP-532,903 is a selective adenosine A3 subtype receptor agonist. It has antiinflammatory effects and has been shown to reduce superoxide generation in damaged tissues,[1] and protects against tissue damage following myocardial ischemia,[2] mediated via an interaction with ATP-sensitive potassium channels.[3]
References
- ↑ van der Hoeven D, Wan TC, Auchampach JA. Activation of the A(3) adenosine receptor suppresses superoxide production and chemotaxis of mouse bone marrow neutrophils. Molecular Pharmacology. 2008 Sep;74(3):685-96. PMID 18583455
- ↑ Tracey WR, Magee WP, Oleynek JJ, Hill RJ, Smith AH, Flynn DM, Knight DR. Novel N6-substituted adenosine 5'-N-methyluronamides with high selectivity for human adenosine A3 receptors reduce ischemic myocardial injury. American Journal of Physiology. Heart and Circulatory Physiology. 2003 Dec;285(6):H2780-7. PMID 12919933
- ↑ Wan TC, Ge ZD, Tampo A, Mio Y, Bienengraeber MW, Tracey WR, Gross GJ, Kwok WM, Auchampach JA. The A3 adenosine receptor agonist CP-532,903 [N6-(2,5-dichlorobenzyl)-3'-aminoadenosine-5'-N-methylcarboxamide] protects against myocardial ischemia/reperfusion injury via the sarcolemmal ATP-sensitive potassium channel. Journal of Pharmacology and Experimental Therapeutics. 2008 Jan;324(1):234-43. doi:10.1124/jpet.107.127480 PMID 17906066
 |

