Biology:Ubiquitin
| Ubiquitin family | |||||||||
|---|---|---|---|---|---|---|---|---|---|
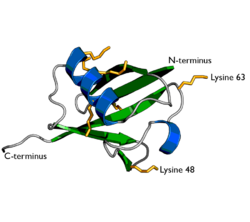 A diagram of ubiquitin. The seven lysine sidechains are shown in yellow/orange. | |||||||||
| Identifiers | |||||||||
| Symbol | ubiquitin | ||||||||
| Pfam | PF00240 | ||||||||
| InterPro | IPR000626 | ||||||||
| PROSITE | PDOC00271 | ||||||||
| SCOP2 | 1aar / SCOPe / SUPFAM | ||||||||
| |||||||||
Ubiquitin is a small (8.6 kDa) regulatory protein found in most tissues of eukaryotic organisms, i.e., it is found ubiquitously. It was discovered in 1975[1] by Gideon Goldstein and further characterized throughout the late 1970s and 1980s.[2] Four genes in the human genome code for ubiquitin: UBB, UBC, UBA52 and RPS27A.[3]
The addition of ubiquitin to a substrate protein is called ubiquitylation (or ubiquitination or ubiquitinylation). Ubiquitylation affects proteins in many ways: it can mark them for degradation via the proteasome, alter their cellular location, affect their activity, and promote or prevent protein interactions.[4][5][6] Ubiquitylation involves three main steps: activation, conjugation, and ligation, performed by ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s), respectively. The result of this sequential cascade is to bind ubiquitin to lysine residues on the protein substrate via an isopeptide bond, cysteine residues through a thioester bond, serine and threonine residues through an ester bond, or the amino group of the protein's N-terminus via a peptide bond.[7][8][9]
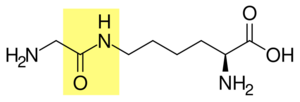
The protein modifications can be either a single ubiquitin protein (monoubiquitylation) or a chain of ubiquitin (polyubiquitylation). Secondary ubiquitin molecules are always linked to one of the seven lysine residues or the N-terminal methionine of the previous ubiquitin molecule. These 'linking' residues are represented by a "K" or "M" (the one-letter amino acid notation of lysine and methionine, respectively) and a number, referring to its position in the ubiquitin molecule as in K48, K29 or M1. The first ubiquitin molecule is covalently bound through its C-terminal carboxylate group to a particular lysine, cysteine, serine, threonine or N-terminus of the target protein. Polyubiquitylation occurs when the C-terminus of another ubiquitin is linked to one of the seven lysine residues or the first methionine on the previously added ubiquitin molecule, creating a chain. This process repeats several times, leading to the addition of several ubiquitins. Only polyubiquitylation on defined lysines, mostly on K48 and K29, is related to degradation by the proteasome (referred to as the "molecular kiss of death"), while other polyubiquitylations (e.g. on K63, K11, K6 and M1) and monoubiquitylations may regulate processes such as endocytic trafficking, inflammation, translation and DNA repair.[10]
The discovery that ubiquitin chains target proteins to the proteasome, which degrades and recycles proteins, was honored with the Nobel Prize in Chemistry in 2004.[8][11][12]
Identification
Ubiquitin (originally, ubiquitous immunopoietic polypeptide) was first identified in 1975[1] as an 8.6 kDa protein expressed in all eukaryotic cells. The basic functions of ubiquitin and the components of the ubiquitylation pathway were elucidated in the early 1980s at the Technion by Aaron Ciechanover, Avram Hershko, and Irwin Rose for which the Nobel Prize in Chemistry was awarded in 2004.[11]
The ubiquitylation system was initially characterised as an ATP-dependent proteolytic system present in cellular extracts. A heat-stable polypeptide present in these extracts, ATP-dependent proteolysis factor 1 (APF-1), was found to become covalently attached to the model protein substrate lysozyme in an ATP- and Mg2+-dependent process.[13] Multiple APF-1 molecules were linked to a single substrate molecule by an isopeptide linkage, and conjugates were found to be rapidly degraded with the release of free APF-1. Soon after APF-1-protein conjugation was characterised, APF-1 was identified as ubiquitin. The carboxyl group of the C-terminal glycine residue of ubiquitin (Gly76) was identified as the moiety conjugated to substrate lysine residues.
The protein
| Number of residues | 76 |
|---|---|
| Molecular mass | 8564.8448 Da |
| Isoelectric point (pI) | 6.79 |
| Gene names | RPS27A (UBA80, UBCEP1), UBA52 (UBCEP2), UBB, UBC |
| Sequence (single-letter) | Template:Monodiv |
Ubiquitin is a small protein that exists in all eukaryotic cells. It performs its myriad functions through conjugation to a large range of target proteins. A variety of different modifications can occur. The ubiquitin protein itself consists of 76 amino acids and has a molecular mass of about 8.6 kDa. Key features include its C-terminal tail and the 7 lysine residues. It is highly conserved throughout eukaryote evolution; human and yeast ubiquitin share 96% sequence identity.[14]
Genes
Ubiquitin is encoded in mammals by 4 different genes. UBA52 and RPS27A genes code for a single copy of ubiquitin fused to the ribosomal proteins L40 and S27a, respectively. The UBB and UBC genes code for polyubiquitin precursor proteins.[3]
Ubiquitylation
Ubiquitylation (also known as ubiquitination or ubiquitinylation) is an enzymatic post-translational modification in which a ubiquitin protein is attached to a substrate protein. This process most commonly binds the last amino acid of ubiquitin (glycine 76) to a lysine residue on the substrate. An isopeptide bond is formed between the carboxyl group (COO−) of the ubiquitin's glycine and the epsilon-amino group (ε-NH+3) of the substrate's lysine.[15] Trypsin cleavage of a ubiquitin-conjugated substrate leaves a di-glycine "remnant" that is used to identify the site of ubiquitylation.[16][17] Ubiquitin can also be bound to other sites in a protein which are electron-rich nucleophiles, termed "non-canonical ubiquitylation".[9] This was first observed with the amine group of a protein's N-terminus being used for ubiquitylation, rather than a lysine residue, in the protein MyoD[18] and has been observed since in 22 other proteins in multiple species,[19][20][21][22][23][24][25][26][27][28][29][30][31][32][33][34][35][36][37] including ubiquitin itself.[38][39] There is also increasing evidence for nonlysine residues as ubiquitylation targets using non-amine groups, such as the sulfhydryl group on cysteine,[34][35][40][41][42][43][44][45][46][47] and the hydroxyl group on threonine and serine.[34][35][40][46][47][48][49][50][51] The end result of this process is the addition of one ubiquitin molecule (monoubiquitylation) or a chain of ubiquitin molecules (polyubiquitination) to the substrate protein.[52]
Ubiquitination requires three types of enzyme: ubiquitin-activating enzymes, ubiquitin-conjugating enzymes, and ubiquitin ligases, known as E1s, E2s, and E3s, respectively. The process consists of three main steps:
- Activation: Ubiquitin is activated in a two-step reaction by an E1 ubiquitin-activating enzyme, which is dependent on ATP. The initial step involves production of a ubiquitin-adenylate intermediate. The E1 binds both ATP and ubiquitin and catalyses the acyl-adenylation of the C-terminus of the ubiquitin molecule. The second step transfers ubiquitin to an active site cysteine residue, with release of AMP. This step results in a thioester linkage between the C-terminal carboxyl group of ubiquitin and the E1 cysteine sulfhydryl group.[15][53] The human genome contains two genes that produce enzymes capable of activating ubiquitin: UBA1 and UBA6.[54]
- Conjugation: E2 ubiquitin-conjugating enzymes catalyse the transfer of ubiquitin from E1 to the active site cysteine of the E2 via a trans(thio)esterification reaction. In order to perform this reaction, the E2 binds to both activated ubiquitin and the E1 enzyme. Humans possess 35 different E2 enzymes, whereas other eukaryotic organisms have between 16 and 35. They are characterised by their highly conserved structure, known as the ubiquitin-conjugating catalytic (UBC) fold.[55]
- Ligation: E3 ubiquitin ligases catalyse the final step of the ubiquitylation cascade. Most commonly, they create an isopeptide bond between a lysine of the target protein and the C-terminal glycine of ubiquitin. In general, this step requires the activity of one of the hundreds of E3s. E3 enzymes function as the substrate recognition modules of the system and are capable of interaction with both E2 and substrate. Some E3 enzymes also activate the E2 enzymes. E3 enzymes possess one of two domains: the homologous to the E6-AP carboxyl terminus (HECT) domain and the really interesting new gene (RING) domain (or the closely related U-box domain). HECT domain E3s transiently bind ubiquitin in this process (an obligate thioester intermediate is formed with the active-site cysteine of the E3), whereas RING domain E3s catalyse the direct transfer from the E2 enzyme to the substrate.[56] The anaphase-promoting complex (APC) and the SCF complex (for Skp1-Cullin-F-box protein complex) are two examples of multi-subunit E3s involved in recognition and ubiquitylation of specific target proteins for degradation by the proteasome.[57]
In the ubiquitylation cascade, E1 can bind with many E2s, which can bind with hundreds of E3s in a hierarchical way. Having levels within the cascade allows tight regulation of the ubiquitylation machinery.[7] Other ubiquitin-like proteins (UBLs) are also modified via the E1–E2–E3 cascade, although variations in these systems do exist.[58]
E4 enzymes, or ubiquitin-chain elongation factors, are capable of adding pre-formed polyubiquitin chains to substrate proteins.[59] For example, multiple monoubiquitylation of the tumor suppressor p53 by Mdm2[60] can be followed by addition of a polyubiquitin chain using p300 and CBP.[61][62]
Types
Ubiquitylation affects cellular process by regulating the degradation of proteins (via the proteasome and lysosome), coordinating the cellular localization of proteins, activating and inactivating proteins, and modulating protein–protein interactions.[4][5][6] These effects are mediated by different types of substrate ubiquitylation, for example the addition of a single ubiquitin molecule (monoubiquitylation) or different types of ubiquitin chains (polyubiquitylation).[63]
Monoubiquitylation
Monoubiquitylation is the addition of one ubiquitin molecule to one substrate protein residue. Multi-monoubiquitylation is the addition of one ubiquitin molecule to multiple substrate residues. The monoubiquitylation of a protein can have different effects to the polyubiquitylation of the same protein. The addition of a single ubiquitin molecule is thought to be required prior to the formation of polyubiquitin chains.[63] Monoubiquitylation affects cellular processes such as membrane trafficking, endocytosis and viral budding.[10][64]
Polyubiquitin chains
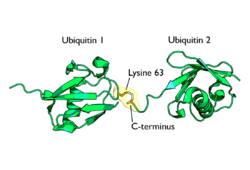
Polyubiquitylation is the formation of a ubiquitin chain on a single lysine residue on the substrate protein. Following addition of a single ubiquitin moiety to a protein substrate, further ubiquitin molecules can be added to the first, yielding a polyubiquitin chain.[63] These chains are made by linking the glycine residue of a ubiquitin molecule to a lysine of ubiquitin bound to a substrate. Ubiquitin has seven lysine residues and an N-terminus that serves as points of ubiquitination; they are K6, K11, K27, K29, K33, K48, K63 and M1, respectively.[8] Lysine 48-linked chains were the first identified and are the best-characterised type of ubiquitin chain. K63 chains have also been well-characterised, whereas the function of other lysine chains, mixed chains, branched chains, M1-linked linear chains, and heterologous chains (mixtures of ubiquitin and other ubiquitin-like proteins) remains more unclear.[17][39][63][64][65]
Lysine 48-linked polyubiquitin chains target proteins for destruction, by a process known as proteolysis. Multi-ubiquitin chains at least four ubiquitin molecules long must be attached to a lysine residue on the condemned protein in order for it to be recognised by the 26S proteasome.[66] This is a barrel-shape structure comprising a central proteolytic core made of four ring structures, flanked by two cylinders that selectively allow entry of ubiquitylated proteins. Once inside, the proteins are rapidly degraded into small peptides (usually 3–25 amino acid residues in length). Ubiquitin molecules are cleaved off the protein immediately prior to destruction and are recycled for further use.[67] Although the majority of protein substrates are ubiquitylated, there are examples of non-ubiquitylated proteins targeted to the proteasome.[68] The polyubiquitin chains are recognised by a subunit of the proteasome: S5a/Rpn10. This is achieved by a ubiquitin-interacting motif (UIM) found in a hydrophobic patch in the C-terminal region of the S5a/Rpn10 unit.[4]
Lysine 63-linked chains are not associated with proteasomal degradation of the substrate protein. Instead, they allow the coordination of other processes such as endocytic trafficking, inflammation, translation, and DNA repair.[10] In cells, lysine 63-linked chains are bound by the ESCRT-0 complex, which prevents their binding to the proteasome. This complex contains two proteins, Hrs and STAM1, that contain a UIM, which allows it to bind to lysine 63-linked chains.[69][70]
Methionine 1-linked (or linear) polyubiquitin chains are another type of non-degradative ubiquitin chains. In this case, ubiquitin is linked in a head-to-tail manner, meaning that the C-terminus of the last ubiquitin molecule binds directly to the N-terminus of the next one. Although initially believed to target proteins for proteasomal degradation,[71] linear ubiquitin later proved to be indispensable for NF-kB signaling.[72] Currently, there is only one known E3 ubiquitin ligase generating M1-linked polyubiquitin chains - linear ubiquitin chain assembly complex (LUBAC).[39][73]
Less is understood about atypical (non-lysine 48-linked) ubiquitin chains but research is starting to suggest roles for these chains.[64] There is evidence that atypical chains linked by lysine 6, 11, 27, 29 and methionine 1 can induce proteasomal degradation.[68][74]
Branched ubiquitin chains containing multiple linkage types can be formed.[75] The function of these chains is unknown.[8]
Structure
Differently linked chains have specific effects on the protein to which they are attached, caused by differences in the conformations of the protein chains. K29-, K33-,[76] K63- and M1-linked chains have a fairly linear conformation; they are known as open-conformation chains. K6-, K11-, and K48-linked chains form closed conformations. The ubiquitin molecules in open-conformation chains do not interact with each other, except for the covalent isopeptide bonds linking them together. In contrast, the closed conformation chains have interfaces with interacting residues. Altering the chain conformations exposes and conceals different parts of the ubiquitin protein, and the different linkages are recognized by proteins that are specific for the unique topologies that are intrinsic to the linkage. Proteins can specifically bind to ubiquitin via ubiquitin-binding domains (UBDs). The distances between individual ubiquitin units in chains differ between lysine 63- and 48-linked chains. The UBDs exploit this by having small spacers between ubiquitin-interacting motifs that bind lysine 48-linked chains (compact ubiquitin chains) and larger spacers for lysine 63-linked chains. The machinery involved in recognising polyubiquitin chains can also differentiate between K63-linked chains and M1-linked chains, demonstrated by the fact that the latter can induce proteasomal degradation of the substrate.[8][10][74]
Function
The ubiquitylation system functions in a wide variety of cellular processes, including:[77]
- Antigen processing
- Apoptosis
- Biogenesis of organelles
- Cell cycle and division
- DNA transcription and repair
- Differentiation and development
- Immune response and inflammation
- Neural and muscular degeneration
- Maintenance of pluripotency[78]
- Morphogenesis of neural networks
- Modulation of cell surface receptors, ion channels and the secretory pathway
- Response to stress and extracellular modulators
- Ribosome biogenesis
- Viral infection
Membrane proteins
Multi-monoubiquitylation can mark transmembrane proteins (for example, receptors) for removal from membranes (internalisation) and fulfil several signalling roles within the cell. When cell-surface transmembrane molecules are tagged with ubiquitin, the subcellular localization of the protein is altered, often targeting the protein for destruction in lysosomes. This serves as a negative feedback mechanism, because often the stimulation of receptors by ligands increases their rate of ubiquitylation and internalisation. Like monoubiquitylation, lysine 63-linked polyubiquitin chains also has a role in the trafficking some membrane proteins.[10][63][66][79]
Genomic maintenance
Proliferating cell nuclear antigen (PCNA) is a protein involved in DNA synthesis. Under normal physiological conditions PCNA is sumoylated (a similar post-translational modification to ubiquitylation). When DNA is damaged by ultra-violet radiation or chemicals, the SUMO molecule that is attached to a lysine residue is replaced by ubiquitin. Monoubiquitylated PCNA recruits polymerases that can carry out DNA synthesis with damaged DNA; but this is very error-prone, possibly resulting in the synthesis of mutated DNA. Lysine 63-linked polyubiquitylation of PCNA allows it to perform a less error-prone mutation bypass known by the template switching pathway.[6][80][81]
Ubiquitylation of histone H2AX is involved in DNA damage recognition of DNA double-strand breaks. Lysine 63-linked polyubiquitin chains are formed on H2AX histone by the E2/E3 ligase pair, Ubc13-Mms2/RNF168.[82][83] This K63 chain appears to recruit RAP80, which contains a UIM, and RAP80 then helps localize BRCA1. This pathway will eventually recruit the necessary proteins for homologous recombination repair.[84]
Transcriptional regulation
Histones can be ubiquitinated, usually in the form of monoubiquitylation, although polyubiquitylated forms do occur. Histone ubiquitylation alters chromatin structure and allows the access of enzymes involved in transcription. Ubiquitin on histones also acts as a binding site for proteins that either activate or inhibit transcription and also can induce further post-translational modifications of the protein. These effects can all modulate the transcription of genes.[85][86]
Deubiquitination
Deubiquitinating enzymes (deubiquitinases; DUBs) oppose the role of ubiquitylation by removing ubiquitin from substrate proteins. They are cysteine proteases that cleave the amide bond between the two proteins. They are highly specific, as are the E3 ligases that attach the ubiquitin, with only a few substrates per enzyme. They can cleave both isopeptide (between ubiquitin and lysine) and peptide bonds (between ubiquitin and the N-terminus). In addition to removing ubiquitin from substrate proteins, DUBs have many other roles within the cell. Ubiquitin is either expressed as multiple copies joined in a chain (polyubiquitin) or attached to ribosomal subunits. DUBs cleave these proteins to produce active ubiquitin. They also recycle ubiquitin that has been bound to small nucleophilic molecules during the ubiquitylation process. Monoubiquitin is formed by DUBs that cleave ubiquitin from free polyubiquitin chains that have been previously removed from proteins.[87][88]
Ubiquitin-binding domains
| Domain | Number of proteins
in proteome |
Length
(amino acids) |
Ubiquitin binding
Affinity |
|---|---|---|---|
| CUE | S. cerevisiae: 7
H. sapiens: 21 |
42–43 | ~2–160 μM |
| GATII | S. cerevisiae: 2
H. sapiens: 14 |
135 | ~180 μM |
| GLUE | S. cerevisiae: ?
H. sapiens: ? |
~135 | ~460 μM |
| NZF | S. cerevisiae: 1
H. sapiens: 25 |
~35 | ~100–400 μM |
| PAZ | S. cerevisiae: 5
H. sapiens: 16 |
~58 | Not known |
| UBA | S. cerevisiae: 10
H. sapiens: 98 |
45–55 | ~0.03–500 μM |
| UEV | S. cerevisiae: 2
H. sapiens: ? |
~145 | ~100–500 μM |
| UIM | S. cerevisiae: 8
H. sapiens: 71 |
~20 | ~100–400 μM |
| VHS | S. cerevisiae: 4
H. sapiens: 28 |
150 | Not known |
Ubiquitin-binding domains (UBDs) are modular protein domains that non-covalently bind to ubiquitin, these motifs control various cellular events. Detailed molecular structures are known for a number of UBDs, binding specificity determines their mechanism of action and regulation, and how it regulates cellular proteins and processes.[89][90]
Disease associations
Pathogenesis
The ubiquitin pathway has been implicated in the pathogenesis of a wide range of diseases and disorders, including:[91]
- Neurodegeneration
- Infection and immunity
- Genetic disorders
- Cancer
Neurodegeneration
Ubiquitin is implicated in neurodegenerative diseases associated with proteostasis dysfunction, including Alzheimer's disease, motor neuron disease,[92] Huntington's disease and Parkinson's disease.[91] Transcript variants encoding different isoforms of ubiquilin-1 are found in lesions associated with Alzheimer's and Parkinson's disease.[93] Higher levels of ubiquilin in the brain have been shown to decrease malformation of amyloid precursor protein (APP), which plays a key role in triggering Alzheimer's disease.[94] Conversely, lower levels of ubiquilin-1 in the brain have been associated with increased malformation of APP.[94] A frameshift mutation in ubiquitin B can result in a truncated peptide missing the C-terminal glycine. This abnormal peptide, known as UBB+1, has been shown to accumulate selectively in Alzheimer's disease and other tauopathies.
Infection and immunity
Ubiquitin and ubiquitin-like molecules extensively regulate immune signal transduction pathways at virtually all stages, including steady-state repression, activation during infection, and attenuation upon clearance. Without this regulation, immune activation against pathogens may be defective, resulting in chronic disease or death. Alternatively, the immune system may become hyperactivated and organs and tissues may be subjected to autoimmune damage.
On the other hand, viruses must block or redirect host cell processes including immunity to effectively replicate, yet many viruses relevant to disease have informationally limited genomes. Because of its very large number of roles in the cell, manipulating the ubiquitin system represents an efficient way for such viruses to block, subvert or redirect critical host cell processes to support their own replication.[95]
The retinoic acid-inducible gene I (RIG-I) protein is a primary immune system sensor for viral and other invasive RNA in human cells.[96] The RIG-I-like receptor (RLR) immune signaling pathway is one of the most extensively studied in terms of the role of ubiquitin in immune regulation.[97]
Genetic disorders
- Angelman syndrome is caused by a disruption of UBE3A, which encodes a ubiquitin ligase (E3) enzyme termed E6-AP.
- Von Hippel–Lindau syndrome involves disruption of a ubiquitin E3 ligase termed the VHL tumor suppressor, or VHL gene.
- Fanconi anemia: Eight of the thirteen identified genes whose disruption can cause this disease encode proteins that form a large ubiquitin ligase (E3) complex.
- 3-M syndrome is an autosomal-recessive growth retardation disorder associated with mutations of the Cullin7 E3 ubiquitin ligase.[98]
Diagnostic use
Immunohistochemistry using antibodies to ubiquitin can identify abnormal accumulations of this protein inside cells, indicating a disease process. These protein accumulations are referred to as inclusion bodies (which is a general term for any microscopically visible collection of abnormal material in a cell). Examples include:
- Neurofibrillary tangles in Alzheimer's disease
- Lewy body in Parkinson's disease
- Pick bodies in Pick's disease
- Inclusions in motor neuron disease and Huntington's disease
- Mallory bodies in alcoholic liver disease
- Rosenthal fibers in astrocytes
Link to cancer
Post-translational modification of proteins is a generally used mechanism in eukaryotic cell signaling.[99] Ubiquitylation, ubiquitin conjugation to proteins, is a crucial process for cell cycle progression and cell proliferation and development. Although ubiquitylation usually serves as a signal for protein degradation through the 26S proteasome, it could also serve for other fundamental cellular processes,[99] in endocytosis,[100] enzymatic activation[101] and DNA repair.[102] Moreover, since ubiquitylation functions to tightly regulate the cellular level of cyclins, its misregulation is expected to have severe impacts. First evidence of the importance of the ubiquitin/proteasome pathway in oncogenic processes was observed due to the high antitumor activity of proteasome inhibitors.[103][104][105] Various studies have shown that defects or alterations in ubiquitylation processes are commonly associated with or present in human carcinoma.[106][107][108][109][110][111][112][113] Malignancies could be developed through loss of function mutation directly at the tumor suppressor gene, increased activity of ubiquitylation, and/or indirect attenuation of ubiquitylation due to mutation in related proteins.[114]
Direct loss of function mutation of E3 ubiquitin ligase
Renal cell carcinoma
The VHL (Von Hippel–Lindau) gene encodes a component of an E3 ubiquitin ligase. VHL complex targets a member of the hypoxia-inducible transcription factor family (HIF) for degradation by interacting with the oxygen-dependent destruction domain under normoxic conditions. HIF activates downstream targets such as the vascular endothelial growth factor (VEGF), promoting angiogenesis. Mutations in VHL prevent degradation of HIF and thus lead to the formation of hypervascular lesions and renal tumors.[106][114]
Breast cancer
The BRCA1 gene is another tumor suppressor gene in humans which encodes the BRCA1 protein that is involved in response to DNA damage. The protein contains a RING motif with E3 Ubiquitin Ligase activity. BRCA1 could form dimer with other molecules, such as BARD1 and BAP1, for its ubiquitylation activity. Mutations that affect the ligase function are often found and associated with various cancers.[110][114]
Cyclin E
As processes in cell cycle progression are the most fundamental processes for cellular growth and differentiation, and are the most common to be altered in human carcinomas, it is expected for cell cycle-regulatory proteins to be under tight regulation. The level of cyclins, as the name suggests, is high only at certain a time point during the cell cycle. This is achieved by continuous control of cyclins or CDKs levels through ubiquitylation and degradation. When cyclin E is partnered with CDK2 and gets phosphorylated, an SCF-associated F-box protein Fbw7 recognizes the complex and thus targets it for degradation. Mutations in Fbw7 have been found in more than 30% of human tumors, characterizing it as a tumor suppressor protein.[113]
Increased ubiquitination activity
Cervical cancer
Oncogenic types of the human papillomavirus (HPV) are known to hijack cellular ubiquitin-proteasome pathway for viral infection and replication. The E6 proteins of HPV will bind to the N-terminus of the cellular E6-AP E3 ubiquitin ligase, redirecting the complex to bind p53, a well-known tumor suppressor gene whose inactivation is found in many types of cancer.[108] Thus, p53 undergoes ubiquitylation and proteasome-mediated degradation. Meanwhile, E7, another one of the early-expressed HPV genes, will bind to Rb, also a tumor suppressor gene, mediating its degradation.[114] The loss of p53 and Rb in cells allows limitless cell proliferation to occur.
p53 regulation
Gene amplification often occur in various tumor cases, including of MDM2, a gene encodes for a RING E3 Ubiquitin ligase responsible for downregulation of p53 activity. MDM2 targets p53 for ubiquitylation and proteasomal degradation thus keeping its level appropriate for normal cell condition. Overexpression of MDM2 causes loss of p53 activity and therefore allowing cells to have a limitless replicative potential.[109][114]
p27
Another gene that is a target of gene amplification is SKP2. SKP2 is an F-box protein with a role in substrate recognition for ubiquitylation and degradation. SKP2 targets p27Kip-1, an inhibitor of cyclin-dependent kinases (CDKs). CDKs2/4 partner with the cyclins E/D, respectively, forming a family of cell cycle regulators which control cell cycle progression through the G1 phase. Low level of p27Kip-1 protein is often found in various cancers and is due to overactivation of ubiquitin-mediated proteolysis through overexpression of SKP2.[111][114]
Efp
Efp, or estrogen-inducible RING-finger protein, is an E3 ubiquitin ligase whose overexpression has been shown to be the major cause of estrogen-independent breast cancer.[105][115] Efp's substrate is 14-3-3 protein which negatively regulates cell cycle.
Evasion of ubiquitination
Colorectal cancer
The gene associated with colorectal cancer is the adenomatous polyposis coli (APC), which is a classic tumor suppressor gene. APC gene product targets beta-catenin for degradation via ubiquitylation at the N-terminus, thus regulating its cellular level. Most colorectal cancer cases are found with mutations in the APC gene. However, in cases where APC gene is not mutated, mutations are found in the N-terminus of beta-catenin which renders it ubiquitination-free and thus increased activity.[107][114]
Glioblastoma
As the most aggressive cancer originated in the brain, mutations found in patients with glioblastoma are related to the deletion of a part of the extracellular domain of the epidermal growth factor receptor (EGFR). This deletion causes CBL E3 ligase unable to bind to the receptor for its recycling and degradation via a ubiquitin-lysosomal pathway. Thus, EGFR is constitutively active in the cell membrane and activates its downstream effectors that are involved in cell proliferation and migration.[112]
Phosphorylation-dependent ubiquitylation
The interplay between ubiquitylation and phosphorylation has been an ongoing research interest since phosphorylation often serves as a marker where ubiquitylation leads to degradation.[99] Moreover, ubiquitylation can also act to turn on/off the kinase activity of a protein.[116] The critical role of phosphorylation is largely underscored in the activation and removal of autoinhibition in the Cbl protein.[117] Cbl is an E3 ubiquitin ligase with a RING finger domain that interacts with its tyrosine kinase binding (TKB) domain, preventing interaction of the RING domain with an E2 ubiquitin-conjugating enzyme. This intramolecular interaction is an autoinhibition regulation that prevents its role as a negative regulator of various growth factors and tyrosine kinase signaling and T-cell activation.[117] Phosphorylation of Y363 relieves the autoinhibition and enhances binding to E2.[117] Mutations that render the Cbl protein dysfunctional due to the loss of its ligase/tumor suppressor function and maintenance of its positive signaling/oncogenic function have been shown to cause the development of cancer.[118][119]
As a drug target
Screening for ubiquitin ligase substrates
Deregulation of E3-substrate interactions is a key cause of many human disorders, therefore identifying E3 ligase substrates is crucial. In 2008, 'Global Protein Stability (GPS) Profiling' was developed to discover E3 ubiquitin ligase substrates.[120] This high-throughput system made use of reporter proteins fused with thousands of potential substrates independently. By inhibition of the ligase activity (through the making of Cul1 dominant negative thus renders ubiquitination not to occur), increased reporter activity shows that the identified substrates are being accumulated. This approach added a large number of new substrates to the list of E3 ligase substrates.
Possible therapeutic applications
Blocking of specific substrate recognition by the E3 ligases, e.g. bortezomib.[115]
Challenge
Finding a specific molecule that selectively inhibits the activity of a certain E3 ligase and/or the protein–protein interactions implicated in the disease remains as one of the important and expanding research area. Moreover, as ubiquitination is a multi-step process with various players and intermediate forms, consideration of the much complex interactions between components needs to be taken heavily into account while designing the small molecule inhibitors.[105]
Similar proteins
Ubiquitin is the most-understood post-translation modifier, however, several family of ubiquitin-like proteins (UBLs) can modify cellular targets in a parallel but distinct route. Known UBLs include: small ubiquitin-like modifier (SUMO), ubiquitin cross-reactive protein (UCRP, also known as interferon-stimulated gene-15 ISG15), ubiquitin-related modifier-1 (URM1), neuronal-precursor-cell-expressed developmentally downregulated protein-8 (NEDD8, also called Rub1 in S. cerevisiae), human leukocyte antigen F-associated (FAT10), autophagy-8 (ATG8) and -12 (ATG12), Few ubiquitin-like protein (FUB1), MUB (membrane-anchored UBL),[121] ubiquitin fold-modifier-1 (UFM1) and ubiquitin-like protein-5 (UBL5, which is but known as homologous to ubiquitin-1 [Hub1] in S. pombe).[122][123] Although these proteins share only modest primary sequence identity with ubiquitin, they are closely related three-dimensionally. For example, SUMO shares only 18% sequence identity, but they contain the same structural fold. This fold is called "ubiquitin fold". FAT10 and UCRP contain two. This compact globular beta-grasp fold is found in ubiquitin, UBLs, and proteins that comprise a ubiquitin-like domain, e.g. the S. cerevisiae spindle pole body duplication protein, Dsk2, and NER protein, Rad23, both contain N-terminal ubiquitin domains.
These related molecules have novel functions and influence diverse biological processes. There is also cross-regulation between the various conjugation pathways, since some proteins can become modified by more than one UBL, and sometimes even at the same lysine residue. For instance, SUMO modification often acts antagonistically to that of ubiquitination and serves to stabilize protein substrates. Proteins conjugated to UBLs are typically not targeted for degradation by the proteasome but rather function in diverse regulatory activities. Attachment of UBLs might, alter substrate conformation, affect the affinity for ligands or other interacting molecules, alter substrate localization, and influence protein stability.
UBLs are structurally similar to ubiquitin and are processed, activated, conjugated, and released from conjugates by enzymatic steps that are similar to the corresponding mechanisms for ubiquitin. UBLs are also translated with C-terminal extensions that are processed to expose the invariant C-terminal LRGG. These modifiers have their own specific E1 (activating), E2 (conjugating) and E3 (ligating) enzymes that conjugate the UBLs to intracellular targets. These conjugates can be reversed by UBL-specific isopeptidases that have similar mechanisms to that of the deubiquitinating enzymes.[77]
Within some species, the recognition and destruction of sperm mitochondria through a mechanism involving ubiquitin is responsible for sperm mitochondria's disposal after fertilization occurs.[124]
Prokaryotic origins
Ubiquitin is believed to have descended from bacterial proteins similar to ThiS (O32583)[125] or MoaD (P30748).[126] These prokaryotic proteins, despite having little sequence identity (ThiS has 14% identity to ubiquitin), share the same protein fold. These proteins also share sulfur chemistry with ubiquitin. MoaD, which is involved in molybdopterin biosynthesis, interacts with MoeB, which acts like an E1 ubiquitin-activating enzyme for MoaD, strengthening the link between these prokaryotic proteins and the ubiquitin system. A similar system exists for ThiS, with its E1-like enzyme ThiF. It is also believed that the Saccharomyces cerevisiae protein Urm1, a ubiquitin-related modifier, is a "molecular fossil" that connects the evolutionary relation with the prokaryotic ubiquitin-like molecules and ubiquitin.[127]
Archaea have a functionally closer homolog of the ubiquitin modification system, where "sampylation" with SAMPs (small archaeal modifier proteins) is performed. The sampylation system only uses E1 to guide proteins to the proteosome.[128] Proteoarchaeota, which are related to the ancestor of eukaryotes, possess all of the E1, E2, and E3 enzymes plus a regulated Rpn11 system. Unlike SAMP which are more similar to ThiS or MoaD, Proteoarchaeota ubiquitin are most similar to eukaryotic homologs.[129]
Prokaryotic ubiquitin-like protein (Pup) and ubiquitin bacterial (UBact)
Prokaryotic ubiquitin-like protein (Pup) is a functional analog of ubiquitin which has been found in the gram-positive bacterial phylum Actinomycetota. It serves the same function (targeting proteins for degradations), although the enzymology of ubiquitylation and pupylation is different, and the two families share no homology. In contrast to the three-step reaction of ubiquitylation, pupylation requires two steps, therefore only two enzymes are involved in pupylation.
In 2017, homologs of Pup were reported in five phyla of gram-negative bacteria, in seven candidate bacterial phyla and in one archaeon[130] The sequences of the Pup homologs are very different from the sequences of Pup in gram-positive bacteria and were termed Ubiquitin bacterial (UBact), although the distinction has yet not been proven to be phylogenetically supported by a separate evolutionary origin and is without experimental evidence.[130]
The finding of the Pup/UBact-proteasome system in both gram-positive and gram-negative bacteria suggests that either the Pup/UBact-proteasome system evolved in bacteria prior to the split into gram positive and negative clades over 3000 million years ago or,[131] that these systems were acquired by different bacterial lineages through horizontal gene transfer(s) from a third, yet unknown, organism. In support of the second possibility, two UBact loci were found in the genome of an uncultured anaerobic methanotrophic Archaeon (ANME-1;locus CBH38808.1 and locus CBH39258.1).
Human proteins containing ubiquitin domain
These include ubiquitin-like proteins.
ANUBL1; BAG1; BAT3/BAG6; C1orf131; DDI1; DDI2; FAU; HERPUD1; HERPUD2; HOPS; IKBKB; ISG15; LOC391257; MIDN; NEDD8; OASL; PARK2; RAD23A; RAD23B; RPS27A; SACS; 8U SF3A1; SUMO1; SUMO2; SUMO3; SUMO4; TMUB1; TMUB2; UBA52; UBB; UBC; UBD; UBFD1; UBL4A; UBL4B; UBL7; UBLCP1; UBQLN1; UBQLN2; UBQLN3; UBQLN4; UBQLNL; UBTD1; UBTD2; UHRF1; UHRF2;
Related proteins
Prediction of ubiquitination
Currently available prediction programs are:
- UbiPred is a SVM-based prediction server using 31 physicochemical properties for predicting ubiquitylation sites.[132]
- UbPred is a random forest-based predictor of potential ubiquitination sites in proteins. It was trained on a combined set of 266 non-redundant experimentally verified ubiquitination sites available from our experiments and from two large-scale proteomics studies.[133]
- CKSAAP_UbSite is SVM-based prediction that employs the composition of k-spaced amino acid pairs surrounding a query site (i.e. any lysine in a query sequence) as input, uses the same dataset as UbPred.[134]
Podcast
Investigating the ubiquitin proteasome system was the focus of a Dementia Researcher Podcast.[135] The podcast was published on 16 August 2021, hosted by Professor Selina Wray from University College London.
See also
- Autophagy
- Autophagin
- Endoplasmic-reticulum-associated protein degradation
- JUNQ and IPOD
- Prokaryotic ubiquitin-like protein
- SUMO enzymes
References
- ↑ 1.0 1.1 "Isolation of a polypeptide that has lymphocyte-differentiating properties and is probably represented universally in living cells". Proceedings of the National Academy of Sciences of the United States of America 72 (1): 11–5. January 1975. doi:10.1073/pnas.72.1.11. PMID 1078892. Bibcode: 1975PNAS...72...11G.
- ↑ "The discovery of ubiquitin-dependent proteolysis". Proceedings of the National Academy of Sciences of the United States of America 102 (43): 15280–2. October 2005. doi:10.1073/pnas.0504842102. PMID 16230621. Bibcode: 2005PNAS..10215280W.
- ↑ 3.0 3.1 "Regulatory mechanisms involved in the control of ubiquitin homeostasis". Journal of Biochemistry 147 (6): 793–8. June 2010. doi:10.1093/jb/mvq044. PMID 20418328.
- ↑ 4.0 4.1 4.2 "The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction". Physiological Reviews 82 (2): 373–428. April 2002. doi:10.1152/physrev.00027.2001. PMID 11917093.
- ↑ 5.0 5.1 "Proteasome-independent functions of ubiquitin in endocytosis and signaling". Science 315 (5809): 201–5. January 2007. doi:10.1126/science.1127085. PMID 17218518. Bibcode: 2007Sci...315..201M.
- ↑ 6.0 6.1 6.2 "Non-traditional functions of ubiquitin and ubiquitin-binding proteins". The Journal of Biological Chemistry 278 (38): 35857–60. September 2003. doi:10.1074/jbc.R300018200. PMID 12860974.
- ↑ 7.0 7.1 "Ubiquitin: structures, functions, mechanisms". Biochimica et Biophysica Acta (BBA) - Molecular Cell Research 1695 (1–3): 55–72. November 2004. doi:10.1016/j.bbamcr.2004.09.019. PMID 15571809.
- ↑ 8.0 8.1 8.2 8.3 8.4 "The ubiquitin code". Annual Review of Biochemistry 81: 203–29. 2012. doi:10.1146/annurev-biochem-060310-170328. PMID 22524316.
- ↑ 9.0 9.1 "Non-canonical ubiquitylation: mechanisms and consequences". The International Journal of Biochemistry & Cell Biology 45 (8): 1833–42. August 2013. doi:10.1016/j.biocel.2013.05.026. PMID 23732108.
- ↑ 10.0 10.1 10.2 10.3 10.4 "Regulation of receptors and transporters by ubiquitination: new insights into surprisingly similar mechanisms". Molecular Interventions 7 (3): 157–67. June 2007. doi:10.1124/mi.7.3.7. PMID 17609522.
- ↑ 11.0 11.1 "The Nobel Prize in Chemistry 2004". Nobelprize.org. http://nobelprize.org/nobel_prizes/chemistry/laureates/2004/.
- ↑ "The Nobel Prize in Chemistry 2004: Popular Information". Nobelprize.org. https://www.nobelprize.org/nobel_prizes/chemistry/laureates/2004/popular.html.
- ↑ "A heat-stable polypeptide component of an ATP-dependent proteolytic system from reticulocytes. 1978". Biochemical and Biophysical Research Communications 425 (3): 565–70. August 2012. doi:10.1016/j.bbrc.2012.08.025. PMID 22925675.
- ↑ Sharp, PM; Li, WH (November 1987). "Molecular evolution of ubiquitin genes.". Trends in ecology & evolution 2 (11): 328-32. doi:10.1016/0169-5347(87)90108-X. PMID 21227875.
- ↑ 15.0 15.1 "Mechanisms underlying ubiquitylation". Annual Review of Biochemistry 70: 503–33. 2001. doi:10.1146/annurev.biochem.70.1.503. PMID 11395416.
- ↑ "Direct identification of a G protein ubiquitylation site by mass spectrometry". Biochemistry 41 (16): 5067–74. April 2002. doi:10.1021/bi015940q. PMID 11955054.
- ↑ 17.0 17.1 "A proteomics approach to understanding protein ubiquitylation". Nature Biotechnology 21 (8): 921–6. August 2003. doi:10.1038/nbt849. PMID 12872131.
- ↑ "A novel site for ubiquitylation: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein". The EMBO Journal 17 (20): 5964–73. October 1998. doi:10.1093/emboj/17.20.5964. PMID 9774340.
- ↑ "Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation". Cell 115 (1): 71–82. October 2003. doi:10.1016/S0092-8674(03)00755-4. PMID 14532004.
- ↑ "The ubiquitin-conjugating enzyme (E2) Ube2w ubiquitinates the N terminus of substrates". The Journal of Biological Chemistry 288 (26): 18784–8. June 2013. doi:10.1074/jbc.C113.477596. PMID 23696636.
- ↑ "The N-terminal domain of MyoD is necessary and sufficient for its nuclear localization-dependent degradation by the ubiquitin system". Proceedings of the National Academy of Sciences of the United States of America 105 (41): 15690–5. October 2008. doi:10.1073/pnas.0808373105. PMID 18836078. Bibcode: 2008PNAS..10515690S.
- ↑ "N-Terminal ubiquitylation of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome". Molecular and Cellular Biology 24 (14): 6140–50. July 2004. doi:10.1128/MCB.24.14.6140-6150.2004. PMID 15226418.
- ↑ "N-terminal polyubiquitylation and degradation of the Arf tumor suppressor". Genes & Development 18 (15): 1862–74. August 2004. doi:10.1101/gad.1213904. PMID 15289458.
- ↑ "The tumor suppressor protein p16(INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitylation at the N-terminal residue". The Journal of Biological Chemistry 279 (40): 41414–21. October 2004. doi:10.1074/jbc.M407201200. PMID 15254040.
- ↑ "Lysine-independent turnover of cyclin G1 can be stabilized by B'alpha subunits of protein phosphatase 2A". Molecular and Cellular Biology 29 (3): 919–28. February 2009. doi:10.1128/MCB.00907-08. PMID 18981217.
- ↑ "Degradation of the E7 human papillomavirus oncoprotein by the ubiquitin-proteasome system: targeting via ubiquitylation of the N-terminal residue". Oncogene 19 (51): 5944–50. November 2000. doi:10.1038/sj.onc.1203989. PMID 11127826.
- ↑ "Degradation of the epstein-barr virus latent membrane protein 1 (LMP1) by the ubiquitin-proteasome pathway. Targeting via ubiquitylation of the N-terminal residue". The Journal of Biological Chemistry 275 (31): 23491–9. August 2000. doi:10.1074/jbc.M002052200. PMID 10807912.
- ↑ "Lysine-independent ubiquitylation of Epstein-Barr virus LMP2A". Virology 300 (1): 153–9. August 2002. doi:10.1006/viro.2002.1562. PMID 12202215.
- ↑ "HSP70 protects BCL2L12 and BCL2L12A from N-terminal ubiquitylation-mediated proteasomal degradation". FEBS Letters 583 (9): 1409–14. May 2009. doi:10.1016/j.febslet.2009.04.011. PMID 19376117.
- ↑ "N-terminal hemagglutinin tag renders lysine-deficient APOBEC3G resistant to HIV-1 Vif-induced degradation by reduced polyubiquitylation". Journal of Virology 85 (9): 4510–9. May 2011. doi:10.1128/JVI.01925-10. PMID 21345952.
- ↑ "Ubiquitin-Proteasome-mediated degradation of Id1 is modulated by MyoD". The Journal of Biological Chemistry 279 (31): 32614–9. July 2004. doi:10.1074/jbc.M403794200. PMID 15163661.
- ↑ "Ubiquitin proteasome-dependent degradation of the transcriptional coactivator PGC-1{alpha} via the N-terminal pathway". The Journal of Biological Chemistry 285 (51): 40192–200. December 2010. doi:10.1074/jbc.M110.131615. PMID 20713359.
- ↑ "Degradation of the Id2 developmental regulator: targeting via N-terminal ubiquitylation". Biochemical and Biophysical Research Communications 314 (2): 505–12. February 2004. doi:10.1016/j.bbrc.2003.12.116. PMID 14733935.
- ↑ 34.0 34.1 34.2 "Ubiquitylation on canonical and non-canonical sites targets the transcription factor neurogenin for ubiquitin-mediated proteolysis". The Journal of Biological Chemistry 284 (23): 15458–68. June 2009. doi:10.1074/jbc.M809366200. PMID 19336407.
- ↑ 35.0 35.1 35.2 "Non-canonical ubiquitylation of the proneural protein Ngn2 occurs in both Xenopus embryos and mammalian cells". Biochemical and Biophysical Research Communications 400 (4): 655–60. October 2010. doi:10.1016/j.bbrc.2010.08.122. PMID 20807509.
- ↑ "Ube2W conjugates ubiquitin to α-amino groups of protein N-termini". The Biochemical Journal 453 (1): 137–45. July 2013. doi:10.1042/BJ20130244. PMID 23560854.
- ↑ "Intrinsic disorder drives N-terminal ubiquitylation by Ube2w". Nature Chemical Biology 11 (1): 83–9. January 2015. doi:10.1038/nchembio.1700. PMID 25436519.
- ↑ "A proteolytic pathway that recognizes ubiquitin as a degradation signal". The Journal of Biological Chemistry 270 (29): 17442–56. July 1995. doi:10.1074/jbc.270.29.17442. PMID 7615550.
- ↑ 39.0 39.1 39.2 "A ubiquitin ligase complex assembles linear polyubiquitin chains". The EMBO Journal 25 (20): 4877–87. October 2006. doi:10.1038/sj.emboj.7601360. PMID 17006537.
- ↑ 40.0 40.1 "Ubiquitination of serine, threonine, or lysine residues on the cytoplasmic tail can induce ERAD of MHC-I by viral E3 ligase mK3". The Journal of Cell Biology 177 (4): 613–24. May 2007. doi:10.1083/jcb.200611063. PMID 17502423.
- ↑ "Ubiquitination on nonlysine residues by a viral E3 ubiquitin ligase". Science 309 (5731): 127–30. July 2005. doi:10.1126/science.1110340. PMID 15994556. Bibcode: 2005Sci...309..127C.
- ↑ "The specificities of Kaposi's sarcoma-associated herpesvirus-encoded E3 ubiquitin ligases are determined by the positions of lysine or cysteine residues within the intracytoplasmic domains of their targets". Journal of Virology 82 (8): 4184–9. April 2008. doi:10.1128/JVI.02264-07. PMID 18272573.
- ↑ "A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p". The Journal of Biological Chemistry 282 (31): 22534–43. August 2007. doi:10.1074/jbc.M702038200. PMID 17550898.
- ↑ "Ubiquitination of mammalian Pex5p, the peroxisomal import receptor". The Journal of Biological Chemistry 282 (43): 31267–72. October 2007. doi:10.1074/jbc.M706325200. PMID 17726030.
- ↑ "A conserved cysteine residue of Pichia pastoris Pex20p is essential for its recycling from the peroxisome to the cytosol". The Journal of Biological Chemistry 282 (10): 7424–30. March 2007. doi:10.1074/jbc.M611627200. PMID 17209040.
- ↑ 46.0 46.1 "Apoptosis induction by Bid requires unconventional ubiquitination and degradation of its N-terminal fragment". The Journal of Cell Biology 179 (7): 1453–66. December 2007. doi:10.1083/jcb.200707063. PMID 18166654.
- ↑ 47.0 47.1 "Complex regulation controls Neurogenin3 proteolysis". Biology Open 1 (12): 1264–72. December 2012. doi:10.1242/bio.20121750. PMID 23259061.
- ↑ "Multilayered mechanism of CD4 downregulation by HIV-1 Vpu involving distinct ER retention and ERAD targeting steps". PLOS Pathogens 6 (4): e1000869. April 2010. doi:10.1371/journal.ppat.1000869. PMID 20442859.
- ↑ "Serine-threonine ubiquitination mediates downregulation of BST-2/tetherin and relief of restricted virion release by HIV-1 Vpu". Journal of Virology 85 (1): 51–63. January 2011. doi:10.1128/JVI.01795-10. PMID 20980512.
- ↑ "Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein". The Journal of Biological Chemistry 285 (31): 23916–24. July 2010. doi:10.1074/jbc.M110.127936. PMID 20519503.
- ↑ "Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids". Molecular Cell 40 (6): 917–26. December 2010. doi:10.1016/j.molcel.2010.11.033. PMID 21172657.
- ↑ "Ubiquitin ligases and beyond". BMC Biology 10: 22. March 2012. doi:10.1186/1741-7007-10-22. PMID 22420755.
- ↑ "Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways". Nature Reviews Molecular Cell Biology 10 (5): 319–31. May 2009. doi:10.1038/nrm2673. PMID 19352404.
- ↑ "Activating the ubiquitin family: UBA6 challenges the field". Trends in Biochemical Sciences 33 (5): 230–7. May 2008. doi:10.1016/j.tibs.2008.01.005. PMID 18353650. http://nbn-resolving.de/urn:nbn:de:bsz:352-opus-85098.
- ↑ "The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins". FASEB Journal 24 (4): 981–93. April 2010. doi:10.1096/fj.09-136259. PMID 19940261.
- ↑ "HECT and RING finger families of E3 ubiquitin ligases at a glance". Journal of Cell Science 125 (Pt 3): 531–7. February 2012. doi:10.1242/jcs.091777. PMID 22389392.
- ↑ "Control of cell growth by the SCF and APC/C ubiquitin ligases". Current Opinion in Cell Biology 21 (6): 816–24. December 2009. doi:10.1016/j.ceb.2009.08.004. PMID 19775879.
- ↑ "Modification of proteins by ubiquitin and ubiquitin-like proteins". Annual Review of Cell and Developmental Biology 22: 159–80. 2006. doi:10.1146/annurev.cellbio.22.010605.093503. PMID 16753028.
- ↑ "A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly". Cell 96 (5): 635–44. March 1999. doi:10.1016/S0092-8674(00)80574-7. PMID 10089879.
- ↑ "Human mdm2 mediates multiple mono-ubiquitination of p53 by a mechanism requiring enzyme isomerization". The Journal of Biological Chemistry 276 (33): 31357–67. August 2001. doi:10.1074/jbc.M011517200. PMID 11397792.
- ↑ "Polyubiquitination of p53 by a ubiquitin ligase activity of p300". Science 300 (5617): 342–4. April 2003. doi:10.1126/science.1080386. PMID 12690203. Bibcode: 2003Sci...300..342G.
- ↑ "CBP and p300 are cytoplasmic E4 polyubiquitin ligases for p53". Proceedings of the National Academy of Sciences of the United States of America 106 (38): 16275–80. September 2009. doi:10.1073/pnas.0904305106. PMID 19805293. Bibcode: 2009PNAS..10616275S.
- ↑ 63.0 63.1 63.2 63.3 63.4 "The emerging complexity of protein ubiquitination". Biochemical Society Transactions 37 (Pt 5): 937–53. October 2009. doi:10.1042/BST0370937. PMID 19754430.
- ↑ 64.0 64.1 64.2 "Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series". EMBO Reports 9 (6): 536–42. June 2008. doi:10.1038/embor.2008.93. PMID 18516089.
- ↑ "Characterization of polyubiquitin chain structure by middle-down mass spectrometry". Analytical Chemistry 80 (9): 3438–44. May 2008. doi:10.1021/ac800016w. PMID 18351785.
- ↑ 66.0 66.1 "Protein regulation by monoubiquitin". Nature Reviews Molecular Cell Biology 2 (3): 195–201. March 2001. doi:10.1038/35056583. PMID 11265249.
- ↑ "Protein degradation by the ubiquitin-proteasome pathway in normal and disease states". Journal of the American Society of Nephrology 17 (7): 1807–19. July 2006. doi:10.1681/ASN.2006010083. PMID 16738015.
- ↑ 68.0 68.1 "Non-canonical ubiquitin-based signals for proteasomal degradation". Journal of Cell Science 125 (Pt 3): 539–48. February 2012. doi:10.1242/jcs.093567. PMID 22389393.
- ↑ "Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes?". The EMBO Journal 32 (4): 552–65. February 2013. doi:10.1038/emboj.2012.354. PMID 23314748.
- ↑ "STAM and Hrs are subunits of a multivalent ubiquitin-binding complex on early endosomes". The Journal of Biological Chemistry 278 (14): 12513–21. April 2003. doi:10.1074/jbc.M210843200. PMID 12551915.
- ↑ Nakamura, Munehiro; Tokunaga, Fuminori; Sakata, Shin-ichi; Iwai, Kazuhiro (December 2006). "Mutual regulation of conventional protein kinase C and a ubiquitin ligase complex" (in en). Biochemical and Biophysical Research Communications 351 (2): 340–347. doi:10.1016/j.bbrc.2006.09.163. PMID 17069764. https://linkinghub.elsevier.com/retrieve/pii/S0006291X06022236.
- ↑ Tokunaga, Fuminori; Sakata, Shin-ichi; Saeki, Yasushi; Satomi, Yoshinori; Kirisako, Takayoshi; Kamei, Kiyoko; Nakagawa, Tomoko; Kato, Michiko et al. (February 2009). "Involvement of linear polyubiquitylation of NEMO in NF-κB activation" (in en). Nature Cell Biology 11 (2): 123–132. doi:10.1038/ncb1821. ISSN 1465-7392. PMID 19136968. http://www.nature.com/articles/ncb1821.
- ↑ Gerlach, Björn; Cordier, Stefanie M.; Schmukle, Anna C.; Emmerich, Christoph H.; Rieser, Eva; Haas, Tobias L.; Webb, Andrew I.; Rickard, James A. et al. (March 2011). "Linear ubiquitination prevents inflammation and regulates immune signalling" (in en). Nature 471 (7340): 591–596. doi:10.1038/nature09816. ISSN 0028-0836. PMID 21455173. Bibcode: 2011Natur.471..591G. http://www.nature.com/articles/nature09816.
- ↑ 74.0 74.1 "Distinct consequences of posttranslational modification by linear versus K63-linked polyubiquitin chains". Proceedings of the National Academy of Sciences of the United States of America 107 (17): 7704–9. April 2010. doi:10.1073/pnas.0908764107. PMID 20385835. Bibcode: 2010PNAS..107.7704Z.
- ↑ "Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages". The Journal of Biological Chemistry 282 (24): 17375–86. June 2007. doi:10.1074/jbc.M609659200. PMID 17426036.
- ↑ "Assembly and specific recognition of k29- and k33-linked polyubiquitin". Molecular Cell 58 (1): 95–109. April 2015. doi:10.1016/j.molcel.2015.01.042. PMID 25752577.
- ↑ 77.0 77.1 "Ubiquitin Proteasome Pathway Overview". http://www.bostonbiochem.com/upp.php.
- ↑ Bax, M (June 2019). "The Ubiquitin Proteasome System Is a Key Regulator of Pluripotent Stem Cell Survival and Motor Neuron Differentiation". Cells 8 (6): 581. doi:10.3390/cells8060581. PMID 31200561.
- ↑ "Deubiquitinase function of A20 maintains and repairs endothelial barrier after lung vascular injury". Cell Death Discovery 4 (60): 60. May 2018. doi:10.1038/s41420-018-0056-3. PMID 29796309.
- ↑ "The Role of PCNA Posttranslational Modifications in Translesion Synthesis". Journal of Nucleic Acids 2010: 1–8. August 2010. doi:10.4061/2010/761217. PMID 20847899.
- ↑ "Regulation of DNA damage responses by ubiquitin and SUMO". Molecular Cell 49 (5): 795–807. March 2013. doi:10.1016/j.molcel.2013.01.017. PMID 23416108.
- ↑ "Molecular insights into the function of RING finger (RNF)-containing proteins hRNF8 and hRNF168 in Ubc13/Mms2-dependent ubiquitylation". The Journal of Biological Chemistry 287 (28): 23900–10. July 2012. doi:10.1074/jbc.M112.359653. PMID 22589545.
- ↑ "DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics". Molecular and Cellular Biology 27 (20): 7028–40. October 2007. doi:10.1128/MCB.00579-07. PMID 17709392.
- ↑ "Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response". Science 316 (5828): 1202–5. May 2007. doi:10.1126/science.1139621. PMID 17525342. Bibcode: 2007Sci...316.1202K.
- ↑ "Ubiquitin-binding domains and their role in the DNA damage response". DNA Repair 8 (4): 544–56. April 2009. doi:10.1016/j.dnarep.2009.01.003. PMID 19213613.
- ↑ "Roles of ubiquitin signaling in transcription regulation". Cellular Signalling 24 (2): 410–21. February 2012. doi:10.1016/j.cellsig.2011.10.009. PMID 22033037.
- ↑ "Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes". Annual Review of Biochemistry 78: 363–97. 2009. doi:10.1146/annurev.biochem.78.082307.091526. PMID 19489724.
- ↑ "A genomic and functional inventory of deubiquitinating enzymes". Cell 123 (5): 773–86. December 2005. doi:10.1016/j.cell.2005.11.007. PMID 16325574.
- ↑ 89.0 89.1 "Ubiquitin-binding domains". Nature Reviews Molecular Cell Biology 6 (8): 610–21. August 2005. doi:10.1038/nrm1701. PMID 16064137.
- ↑ "Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions". Annual Review of Biochemistry 81: 291–322. 2012-01-01. doi:10.1146/annurev-biochem-051810-094654. PMID 22482907.
- ↑ 91.0 91.1 Popovic, D (November 2014). "Ubiquitination in disease pathogenesis and treatment". Nature Medicine 20 (11): 1242–1253. doi:10.1038/nm.3739. PMID 25375928. https://www.nature.com/articles/nm.3739#Abs2.
- ↑ Yerbury, Justin (May 2020). "Proteome Homeostasis Dysfunction: A Unifying Principle in ALS Pathogenesis". Trends in Neurosciences 43 (5): 274‐284. doi:10.1016/j.tins.2020.03.002. PMID 32353332. https://pubmed.ncbi.nlm.nih.gov/32353332/?from_single_result=Proteome+Homeostasis+Dysfunction%3A+A+Unifying+Principle+in+ALS+Pathogenesis.
- ↑ "UBQLN1 ubiquilin 1 [ Homo sapiens "]. Gene. National Center for Biotechnology Information. https://www.ncbi.nlm.nih.gov/sites/entrez?Db=gene&Cmd=DetailsSearch&Term=29979%5Buid%5D.
- ↑ 94.0 94.1 "Ubiquilin-1 is a molecular chaperone for the amyloid precursor protein". The Journal of Biological Chemistry 286 (41): 35689–98. October 2011. doi:10.1074/jbc.M111.243147. PMID 21852239.
- "Alzheimer's brains found to have lower levels of key protein". ScienceDaily (Press release). September 1, 2011.
- ↑ "Ubiquitin in the activation and attenuation of innate antiviral immunity". The Journal of Experimental Medicine 213 (1): 1–13. January 2016. doi:10.1084/jem.20151531. PMID 26712804.
- ↑ "Pattern recognition receptors and inflammation" (in en). Cell 140 (6): 805–20. March 2010. doi:10.1016/j.cell.2010.01.022. PMID 20303872.
- ↑ "Regulation of RIG-I Activation by K63-Linked Polyubiquitination" (in en). Frontiers in Immunology 8: 1942. 2018. doi:10.3389/fimmu.2017.01942. PMID 29354136.
- ↑ "Identification of mutations in CUL7 in 3-M syndrome". Nature Genetics 37 (10): 1119–24. October 2005. doi:10.1038/ng1628. PMID 16142236.
- ↑ 99.0 99.1 99.2 "When ubiquitination meets phosphorylation: a systems biology perspective of EGFR/MAPK signalling". Cell Communication and Signaling 11: 52. July 2013. doi:10.1186/1478-811X-11-52. PMID 23902637.
- ↑ "Endocytosis and intracellular trafficking of ErbBs". Experimental Cell Research 314 (17): 3093–106. October 2008. doi:10.1016/j.yexcr.2008.07.029. PMID 18793634.
- ↑ "Switches, excitable responses and oscillations in the Ring1B/Bmi1 ubiquitination system". PLOS Computational Biology 7 (12): e1002317. December 2011. doi:10.1371/journal.pcbi.1002317. PMID 22194680. Bibcode: 2011PLSCB...7E2317N.
- ↑ "Histone H2A ubiquitination in transcriptional regulation and DNA damage repair". The International Journal of Biochemistry & Cell Biology 41 (1): 12–5. January 2009. doi:10.1016/j.biocel.2008.09.016. PMID 18929679.
- ↑ "Proteasome inhibitors as potential novel anticancer agents". Drug Resistance Updates 2 (4): 215–223. August 1999. doi:10.1054/drup.1999.0095. PMID 11504494.
- ↑ "Clinical Cancer Research 2000: New Agents and Therapies". Drug Resistance Updates 3 (4): 197–201. 2000. doi:10.1054/drup.2000.0153. PMID 11498385.
- ↑ 105.0 105.1 105.2 "Cell cycle regulatory E3 ubiquitin ligases as anticancer targets". Drug Resistance Updates 5 (6): 249–58. December 2002. doi:10.1016/s1368-7646(02)00121-8. PMID 12531181.
- ↑ 106.0 106.1 "Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel–Lindau disease". Human Molecular Genetics 10 (10): 1029–38. 2001. doi:10.1093/hmg/10.10.1029. PMID 11331613.
- ↑ 107.0 107.1 "Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer". Cancer Research 58 (6): 1130–4. March 1998. PMID 9515795.
- ↑ 108.0 108.1 "The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53". Cell 75 (3): 495–505. November 1993. doi:10.1016/0092-8674(93)90384-3. PMID 8221889.
- ↑ 109.0 109.1 "The MDM2 gene amplification database". Nucleic Acids Research 26 (15): 3453–9. August 1998. doi:10.1093/nar/26.15.3453. PMID 9671804.
- ↑ 110.0 110.1 "The RING heterodimer BRCA1-BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation". The Journal of Biological Chemistry 276 (18): 14537–40. May 2001. doi:10.1074/jbc.C000881200. PMID 11278247.
- ↑ 111.0 111.1 "Skp2 gene copy number aberrations are common in non-small cell lung carcinoma, and its overexpression in tumors with ras mutation is a poor prognostic marker". Clinical Cancer Research 10 (6): 1984–91. 2004. doi:10.1158/1078-0432.ccr-03-0470. PMID 15041716.
- ↑ 112.0 112.1 "Epidermal growth factor receptor signaling intensity determines intracellular protein interactions, ubiquitination, and internalization". Proceedings of the National Academy of Sciences of the United States of America 100 (11): 6505–10. May 2003. doi:10.1073/pnas.1031790100. PMID 12734385. Bibcode: 2003PNAS..100.6505S.
- ↑ 113.0 113.1 "DNA copy number losses in human neoplasms". The American Journal of Pathology 155 (3): 683–94. September 1999. doi:10.1016/S0002-9440(10)65166-8. PMID 10487825.
- ↑ 114.0 114.1 114.2 114.3 114.4 114.5 114.6 "The ubiquitin-proteasome pathway and its role in cancer". Journal of Clinical Oncology 23 (21): 4776–89. July 2005. doi:10.1200/JCO.2005.05.081. PMID 16034054.
- ↑ 115.0 115.1 "Therapeutic anti-cancer targets upstream of the proteasome". Cancer Treatment Reviews 29 (Suppl 1): 49–57. May 2003. doi:10.1016/s0305-7372(03)00083-5. PMID 12738243.
- ↑ "Ubiquitylation of MEKK1 inhibits its phosphorylation of MKK1 and MKK4 and activation of the ERK1/2 and JNK pathways". The Journal of Biological Chemistry 278 (3): 1403–6. January 2003. doi:10.1074/jbc.C200616200. PMID 12456688.
- ↑ 117.0 117.1 117.2 "Autoinhibition and phosphorylation-induced activation mechanisms of human cancer and autoimmune disease-related E3 protein Cbl-b". Proceedings of the National Academy of Sciences of the United States of America 108 (51): 20579–84. December 2011. doi:10.1073/pnas.1110712108. PMID 22158902. Bibcode: 2011PNAS..10820579K.
- ↑ "Germline CBL mutations cause developmental abnormalities and predispose to juvenile myelomonocytic leukemia". Nature Genetics 42 (9): 794–800. September 2010. doi:10.1038/ng.641. PMID 20694012.
- ↑ "Cbl and human myeloid neoplasms: the Cbl oncogene comes of age". Cancer Research 70 (12): 4789–94. June 2010. doi:10.1158/0008-5472.CAN-10-0610. PMID 20501843.
- ↑ "Identification of SCF ubiquitin ligase substrates by global protein stability profiling". Science 322 (5903): 923–9. 2008. doi:10.1126/science.1160462. PMID 18988848. Bibcode: 2008Sci...322..923Y.
- ↑ "MUBs, a family of ubiquitin-fold proteins that are plasma membrane-anchored by prenylation". The Journal of Biological Chemistry 281 (37): 27145–57. September 2006. doi:10.1074/jbc.M602283200. PMID 16831869.
- ↑ "Ubiquitin and ubiquitin-like proteins as multifunctional signals". Nature Reviews Molecular Cell Biology 6 (8): 599–609. August 2005. doi:10.1038/nrm1700. PMID 16064136.
- ↑ "Functional roles of ubiquitin-like domain (ULD) and ubiquitin-binding domain (UBD) containing proteins". Chemical Reviews 109 (4): 1481–94. April 2009. doi:10.1021/cr800413p. PMID 19253967.
- ↑ "Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos". Biology of Reproduction 63 (2): 582–90. August 2000. doi:10.1095/biolreprod63.2.582. PMID 10906068.
- ↑ "Solution structure of ThiS and implications for the evolutionary roots of ubiquitin". Nature Structural Biology 8 (1): 47–51. January 2001. doi:10.1038/83041. PMID 11135670.
- ↑ "Mechanism of ubiquitin activation revealed by the structure of a bacterial MoeB-MoaD complex". Nature 414 (6861): 325–9. November 2001. doi:10.1038/35104586. PMID 11713534. Bibcode: 2001Natur.414..325L.
- ↑ "Origin and function of ubiquitin-like proteins". Nature 458 (7237): 422–9. March 2009. doi:10.1038/nature07958. PMID 19325621. Bibcode: 2009Natur.458..422H.
- ↑ "Archaeal Proteasomes and Sampylation". Regulated Proteolysis in Microorganisms. Subcellular Biochemistry. 66. 2013. pp. 297–327. doi:10.1007/978-94-007-5940-4_11. ISBN 978-94-007-5939-8.
- ↑ "Rpn11-mediated ubiquitin processing in an ancestral archaeal ubiquitination system". Nature Communications 9 (1): 2696. July 2018. doi:10.1038/s41467-018-05198-1. PMID 30002364. Bibcode: 2018NatCo...9.2696F.
- ↑ 130.0 130.1 "Identification of UBact, a ubiquitin-like protein, along with other homologous components of a conjugation system and the proteasome in different gram-negative bacteria". Biochemical and Biophysical Research Communications 483 (3): 946–950. February 2017. doi:10.1016/j.bbrc.2017.01.037. PMID 28087277.
- ↑ "The Timetree of Prokaryotes: New Insights into Their Evolution and Speciation". Molecular Biology and Evolution 34 (2): 437–446. February 2017. doi:10.1093/molbev/msw245. PMID 27965376.
- ↑ "Computational identification of ubiquitylation sites from protein sequences". BMC Bioinformatics 9: 310. July 2008. doi:10.1186/1471-2105-9-310. PMID 18625080.
- ↑ "Identification, analysis, and prediction of protein ubiquitination sites". Proteins 78 (2): 365–80. February 2010. doi:10.1002/prot.22555. PMID 19722269.
- ↑ Fraternali, Franca, ed (2011). "Prediction of ubiquitination sites by using the composition of k-spaced amino acid pairs". PLOS ONE 6 (7): e22930. doi:10.1371/journal.pone.0022930. PMID 21829559. Bibcode: 2011PLoSO...622930C.
- ↑ "Stream episode Investigating the ubiquitin proteasome system by Dementia Researcher podcast | Listen online for free on SoundCloud". https://soundcloud.com/dementia-researcher/investigating-the-ubiquitin-proteasome-system.
External links
- GeneReviews/NCBI/NIH/UW entry on Angelman syndrome
- OMIM entries on Angelman syndrome
- UniProt entry for ubiquitin
- "7.340 Ubiquitination: The Proteasome and Human Disease". MIT OpenCourseWare. 2004. http://ocw.mit.edu/courses/biology/7-340-ubiquitination-the-proteasome-and-human-disease-fall-2004/. Notes from MIT course.
- Ubiquitin at the US National Library of Medicine Medical Subject Headings (MeSH)
 |